Fig. 3.—
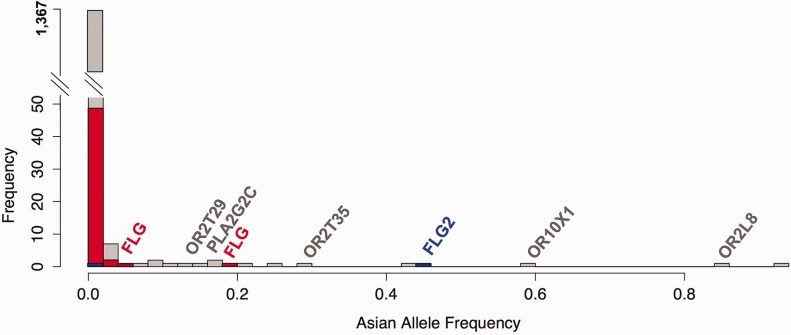
(C) The histogram for the frequencies of multiple FLG loss-of-function variants in the genome is significantly higher than chromosome-wide expectations. The x-axis of this histogram shows the allele frequency of observed loss-of-function variants in Asian population across chromosome 1. The y-axis shows the number of observations. The majority of loss-of-function variants are rare (<1%). However, four different FLG loss-of-function variants are individually have higher allele frequencies, and cumulatively the allele frequency of FLG loss-of-function variants represents one of the highest in the genome.