Figure 4. Class prediction accuracy using leave-one-out cross-validation and weighted gene-voting.
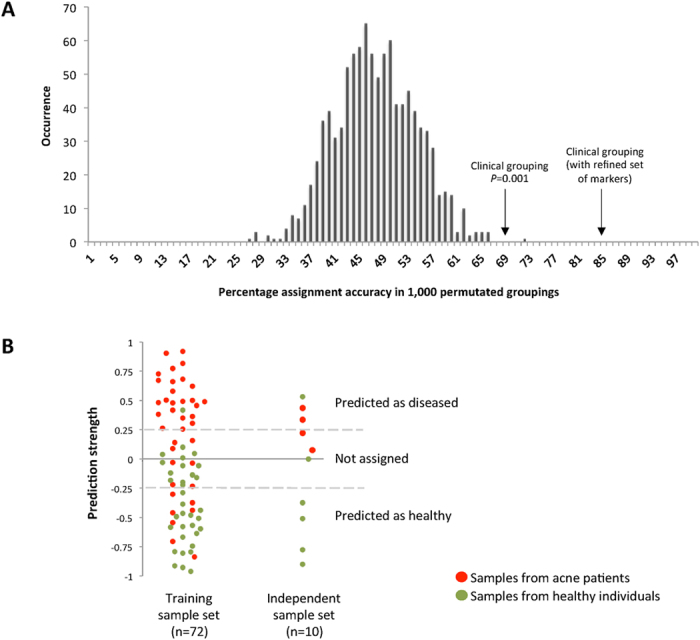
(A) For the training sample set (n = 72), using the clinically defined acne and healthy individual grouping, the classifier correctly assigned the clinical states of 34 of the 49 assigned samples (69% accuracy) using a prediction strength threshold of 0.25. This result is statistically significant (P = 0.001), because only one of the 1,000 permutated groupings had a higher accuracy. However, that particular grouping (accuracy of 72%) had fewer samples assigned than the clinical grouping (n = 39 vs 49). This demonstrates that the differences in the relative abundances of the metagenomic elements between acne patients and healthy individuals can be used to predict the clinical states of the skin. When we used the refined set of 45 metagenomic elements, including 43 P. acnes OGUs, P. acnes locus 2, and P. granulosum, we further improved the prediction accuracy of the training sample set to 85%. (B) Consistent with the prediction accuracy on the training sample set, for the independent sample set (n = 10), the 45 metagenomic elements were able to assign 70% of the samples with 86% accuracy.
