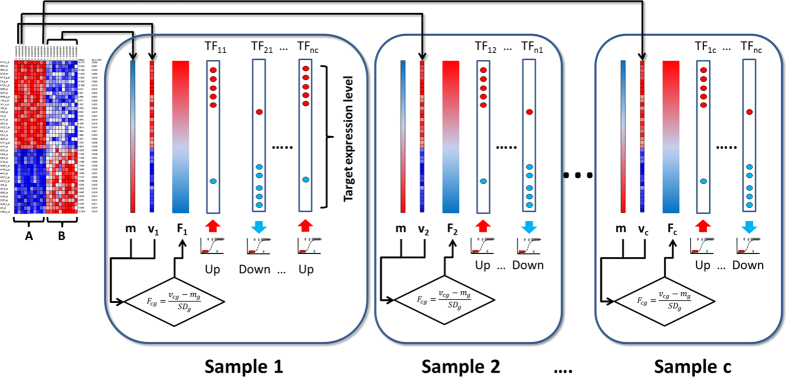
Figure 7. Schema of the TFTEA method to obtain personalized values of survival.
The method uses gene expression values and compares two conditions (A and B). However, in this case all the samples in the (B) condition are used to produce an average expression value for any of the genes in this condition (m vector). Then, each sample in the (A) condition (vc) can be compared to the average expression in the (B) condition and a rank of fold change (Fcg) is generated for each sample. Then, this ranking is used in the same way that the rank of differential expression was used in Fig. 6 to find differentially activated TFs in samples from the condition (A) with respect to the average (B) condition. Is samples are paired the Fcg value can be derived from the direct comparison between them.