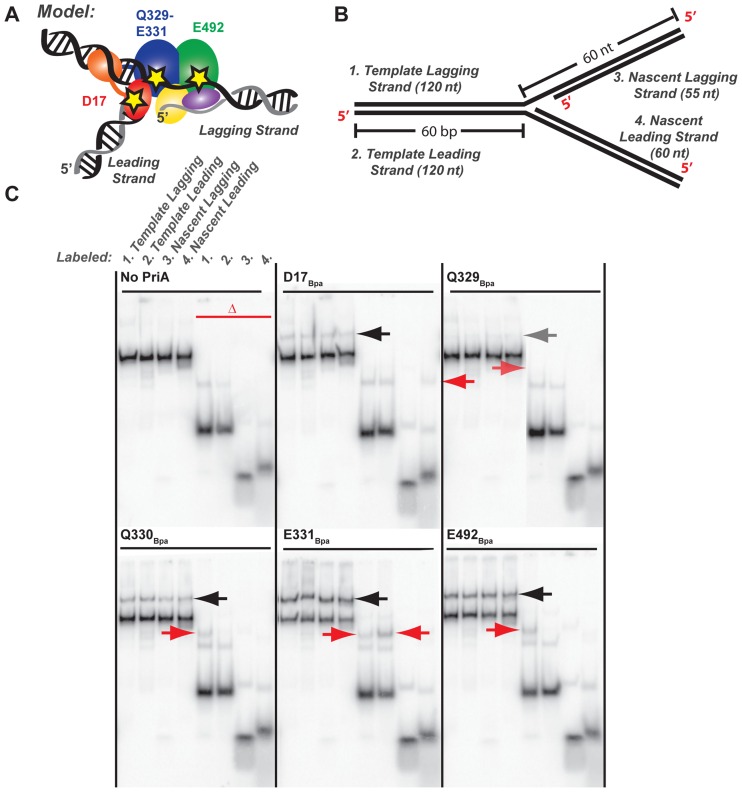
Figure 4.
PriA–DNA fork crosslinking maps strand- and residue-specific interactions. (A) Model of PriA domains predicted to bind to specific DNA regions of an abandoned DNA fork (23). Stars note the location of Bpa incorporation: D17 within the 3′BD, Q329-E331 within the ARL of helicase lobe 1, and E492 within the helicase lobe 2. (B) Synthetic DNA fork substrate used in crosslinking experiments. The substrate is lengthened relative to that used in Figure 3 but it retains the 5 nt nt ssDNA gap between parental duplex and nascent lagging strand 5′ end (Supplementary Table S1 structure #3). (C) Denaturing PAGE separation of DNA-bound, Bpa-incorporated PriA variants after exposure to ultraviolet light. DNA forked substrate in B was individually 5′-radiolabeled on each of the 4 strands, yielding the set of 4 adjacent samples. Half of each sample was additionally heat-denatured, yielding the second set of 4 adjacent samples marked by the red delta. Arrows point to covalently-crosslinked PriA-shifted bands. Data are representative gels from among three replicates.