Figure 5.
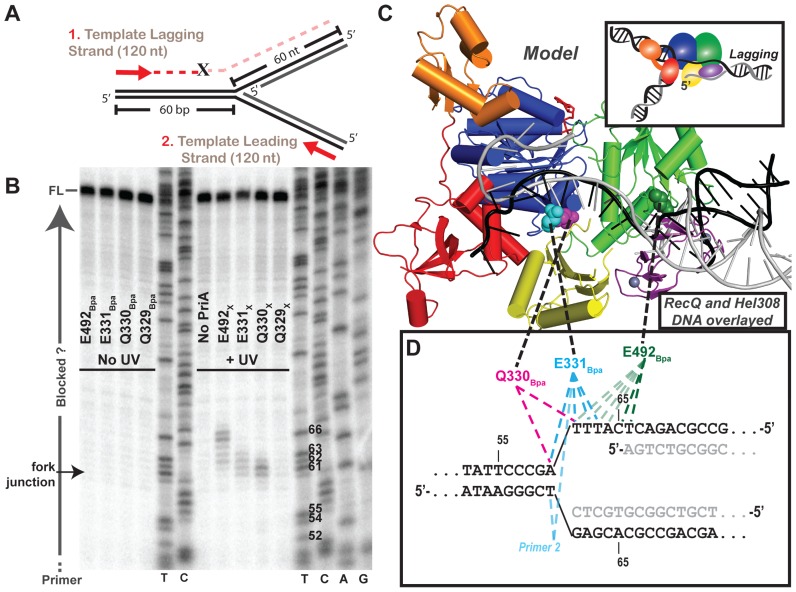
Primer extension maps PriABpa–DNA fork crosslink sites at the nucleotide-level. (A) Diagram of primer extension assay on the crosslinking DNA fork (Figure 4B and Supplementary Table S1 structure #3). Red arrows represent 5′ 32P-labelled primers that allow extension along the template lagging (primer 1) or leading strands (primer 2; Supplementary Table S1). ‘X’ indicates possible location of block to primer extension by a covalent PriA–DNA crosslink. (B) Lagging strand PE products resolved by urea-denaturing PAGE with DNA sequencing ladders in middle and right. T,C,A or G at bottom of sequencing ladder lane denote halted base identity in primer extension product sequence, Thymidine nt # 52–66 are labeled on gel, to right of T lane, corresponding to numbering in panel D. Labeling to left of gel indicates full-length (FL) product, fork junction location and primers (run off). Data are representative gel of greater than three replicates. (C) Overlay of helicase domains from: PriA (multi-coloring as in Figure 1, (23)) and partial duplex DNA-bound RecQ (PDB 4TMU (23)) and Hel308 (PDB 2P6R (45)) structures, showing just the DNA from RecQ structure (black) and Hel308 structure (grey). (D) Sequence of the crosslinking DNA fork at the fork junction with primer extension truncated products for each variant mapped onto DNA fork substrate. Dotted lines connect the structural location of the Bpa-substituted residue in panel C with the mapped nucleotide crosslink location in panel D. As indicated earlier, the DNA includes a 5 nt ssDNA gap present between the parental duplex and nascent lagging strand 5′ end.