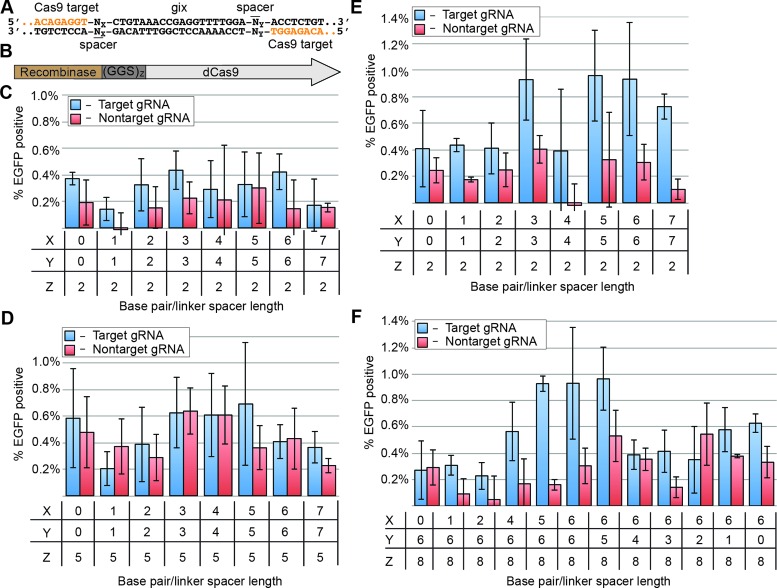
Figure 2.
Optimization of fusion linker lengths and target site spacer variants. A single target guide RNA expression vector, pHU6-NT1 or non-target vector pHU6-BC74 was used in these experiments. See supporting material, Tables S6–S8, for sequences. (A) A portion of the target site is shown with guide RNA target sites in orange and a gix pseudo site in black. The 5′ and 3′ sequences on either side of the pseudo-gix sites are identical, but inverted, and are recognized by pHU6-NT1. The number of base pair spacers separating the gix pseudo-site from the 5′ and 3′ binding sites is represented by an X and Y, respectively. (B) Z represents the number of GGS repeats connecting Ginβ to dCas9. Fusion protein activity is assessed with X = Y for (C) (GGS)2 (D) (GGS)5 and (E) (GGS)8 linkers (i.e. recCas9) connecting the Gin catalytic domain to the dCas9 domain. (F) The activity of recCas9 on target sites composed of uneven base pair spacers (X≠Y) was determined; X = Y = 6 is included for comparison. All experiments are performed in triplicate and background fluorescence is subtracted from these experiments (see Materials and Methods). The percentage of EGFP-positive cells shown is of transfected cells (determined by gating for the presence of a cotransfected plasmid constitutively expressing iRFP) and at least 6000 live events are recorded for each experiment. Guide RNA expression vectors and guide RNA sequences are abbreviated as gRNA. Values and error bars represent the mean and standard deviation, respectively, of three independent biological replicates.