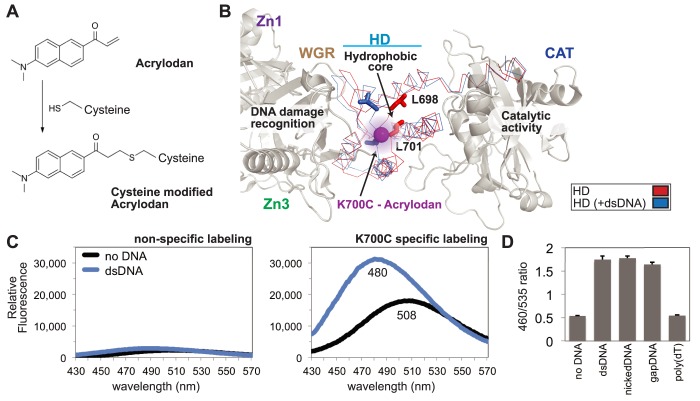
Figure 2.
A PARP-1 sensor to monitor HD destabilization. (A) The small molecule acrylodan reacts specifically with cysteine residues forming a covalent attachment. Upon reaction, this molecule has varied fluorescent peak emission dependent upon local solvent polarity. (B) Structure of PARP-1 essential domains bound to DNA damage (PDB code 4DQY; gray), highlighting and comparing activated HD (PDB code 4DQY; blue) with non-activated HD (PDB code 1A26; red). The major observable difference in the crystal structure lies within the ‘leucine switch’, exemplified by L698 and L701 transitioning out of the HD hydrophic interior. The mutated residue K700C was selectively labeled with acrylodan (magenta sphere), allowing for real-time detection of HD destabilization. (C) Emission profile of acrylodan-labeled PARP-1 in the absence and presence of dsDNA. A construct lacking the engineered cysteine at position 700 (WT_2XC, two native cysteines removed; see Methods) displayed very low fluorescence intensity, indicating that the K700C mutant was selectively labeled. (D) Acrylodan-PARP-1 was used to show specificity for DNA structure by comparing changes in the emission ratio (λ460/λ535) of dsDNA, nickDNA, gapDNA, and poly(dT). For the no DNA control, only the DNA annealing buffer was added. Data points were conducted in triplicates ± S.D.