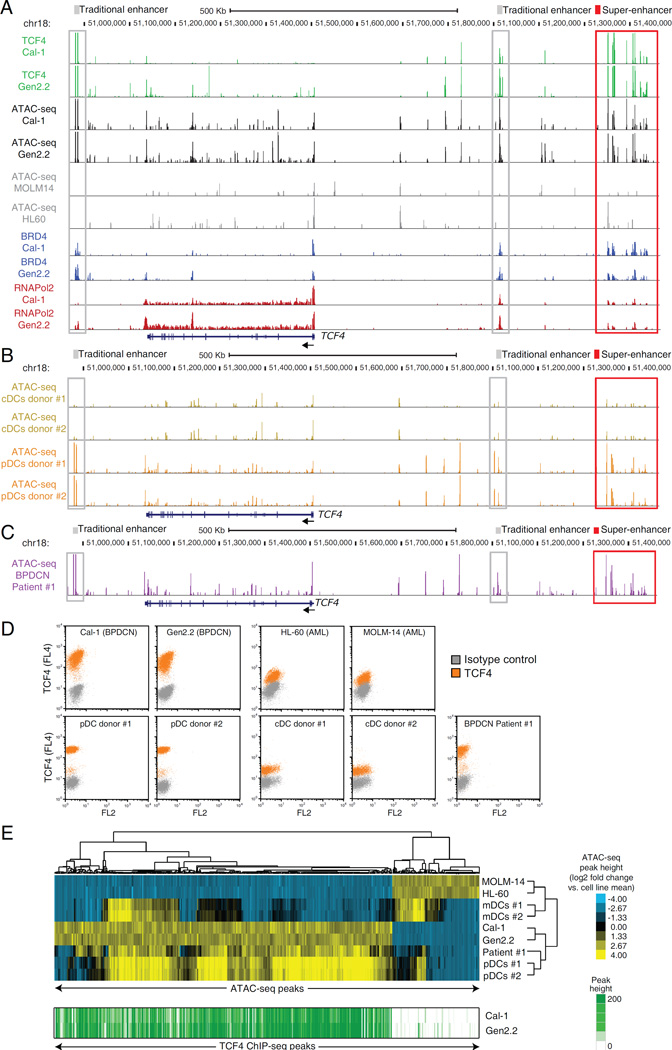
Figure 8.
ATAC-Seq mapping of the regulatory landscape of BPDCN. A) ATAC-Seq was used to map the open chromatin landscape of the TCF4 locus in BPDCN cell lines (black) and in control AML cell lines (gray). TCF4, BRD4 and RNA Pol2 ChIP-Seq in the BPDCN cell lines are also shown in green, blue and red, respectively. B) Normal pDCs and CD1c+ cDCs were isolated from 2 healthy donors .The regulatory landscape of the TCF4 locus is shown in orange and yellow for pDCs and cDCs, respectively. C) ATAC-Seq was used to map the open chromatin regulatory landscape of the TCF4 locus in PBMCs isolated from a patient with leukemic BPDCN (purple). D) TCF4 intracellular flow was performed for the indicated cell lines and primary samples. E) Heat-map results of the unsupervised hierarchical clustering based on the ATAC-Seq predictor regions. Cell lines and primary samples are indicated on the right dendrogram. The corresponding TCF4 ChIP-Seq density is shown in green for the two BPDCN cell lines. See also Figure S8 and Table S8.