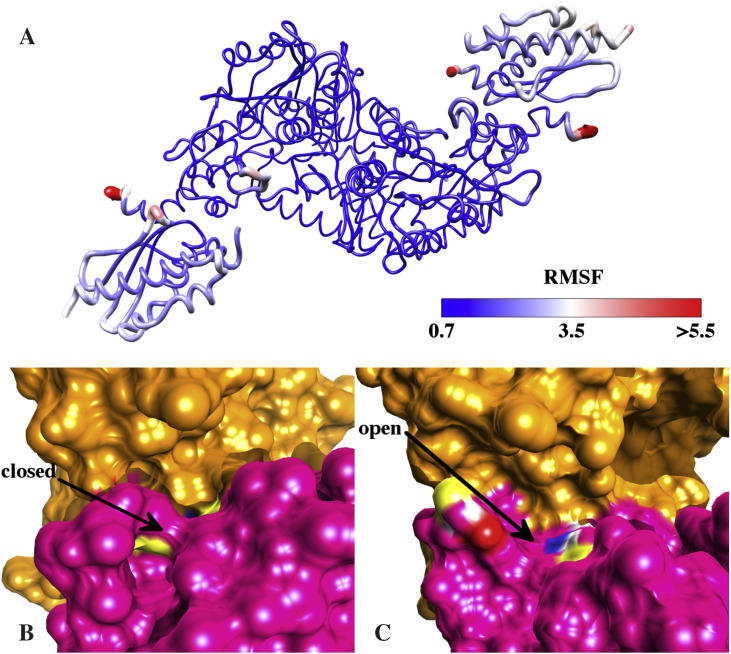
Fig. 2.
Backbone RMSF and IscUβ-turn opening motion. (A) Average structure of the IscS-IscU tetramer and E. coli IscS-IscU crystal structure (PDB ID: 3LVL) from the binary complex simulation, depicted as ribbons and colored according to backbone RMSF values, from lowest (blue) to highest (red). Ribbon thickness is proportional to the local RMSF value. Detailed view of the IscU active site in the binary complex showing a (B) closed form of its β-turn from the E. coli IscS-IscU crystal structure (ID PDB: 3LVL) and (C) an open form of the same turn from the average IscS-IscU complex structure from our simulation. In both B and C panels, IscS (orange) and IscU (pink) are displayed as surfaces.