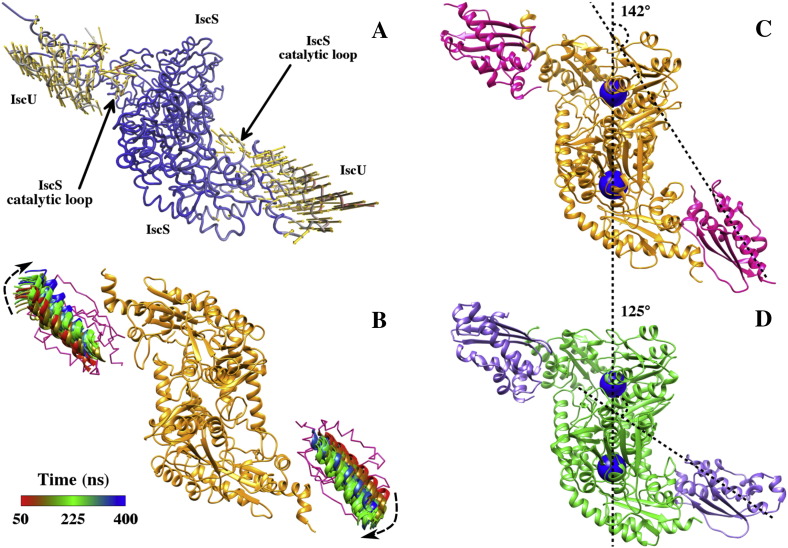
Fig. 4.
IscU orientation and dynamics within the binary complex. (A) Main collective motions of the IscS-IscU complex from PCA (Cα atoms only), showing the IscU pivotal movement around the IscS dimer and the motion of the IscS catalytic loop. Vectors display the amount and direction of the residue motion along the first eigenvector. Minor residue contributions (i.e. arrow length < 2Å) were omitted. (B) Motion of the IscU protomers around the IscS dimer. A number of uncorrelated IscU configurations (one every 10 ns) are superposed upon fitting onto the IscS dimer structure (orange). A representative α-helix of IscU is depicted as cartoons and colored as a function of simulation time, while the rest of the protein is depicted as pink ribbons. Angle formed between IscU and IscS from (C) the average IscS-IscU complex structure from our MD simulation, and (D) the A. fulgidus crystal structure.