Figure 7.
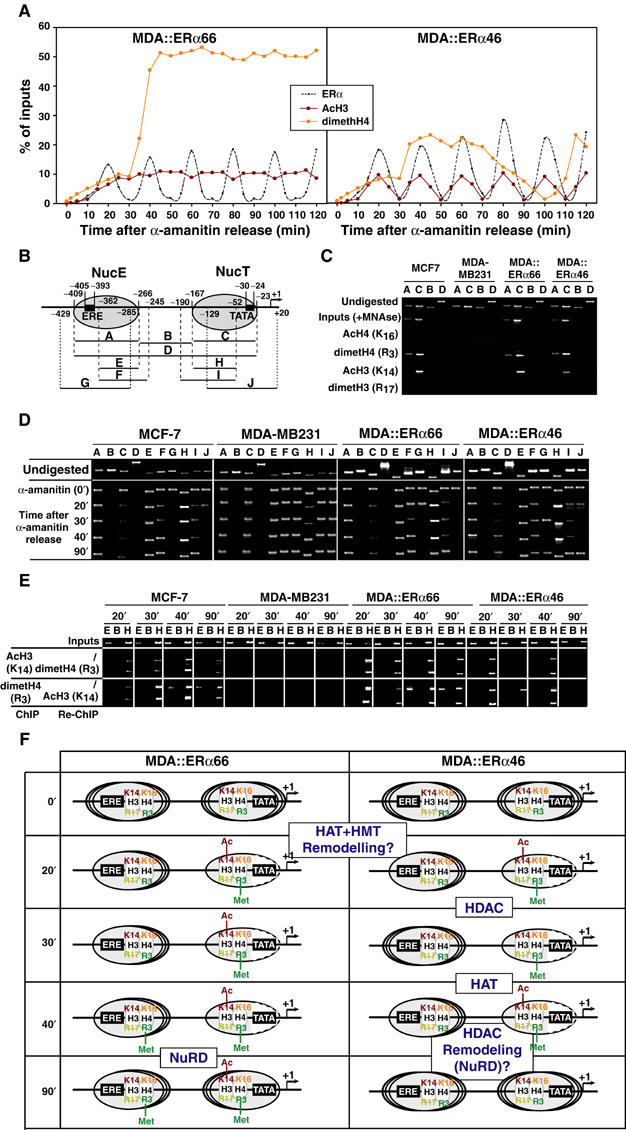
Dynamic chromatin modifications within the pS2 promoter. (A) Kinetic ChIP experiments were performed as depicted in Figure 4, using antibodies and chromatin prepared from synchronized MDA-MB231 cells expressing either ERα isoforms, as indicated within the graphs. (B) Schematic representation of the pS2 promoter, with phased nucleosomes and regions amplified by PCR primers depicted. (C) Mononucleosomes prepared from indicated cells were used in ChIP assays detecting modified histones in both NucE and NucT. The absence of amplification using the B and D primer pairs controls proper digestion. (D) Fluctuations in NucE and NucT positions on the pS2 promoter were assessed through PCR amplification of mononucleosomes using the A to J primer pairs. (E) Following α-amanitin synchronization, mononucleosomes were prepared at times indicated and subjected to ChIP and Re-ChIP procedures using the indicated antibodies. Use of the E and H primer pairs for DNA amplification showed the localization of the modified histones within either NucE or NucT. (F) Scheme summarizing the dynamic histone modifications and nucleosomal organization of the pS2 promoter during ERα isoforms cycles. Dashed lines in NucT reflect the increased accessibility of the TATA box surrounding region for MNase digestion. The recruitment events that are hypothesized to correlate with the observed nucleosome modifications are boxed and highlighted.
