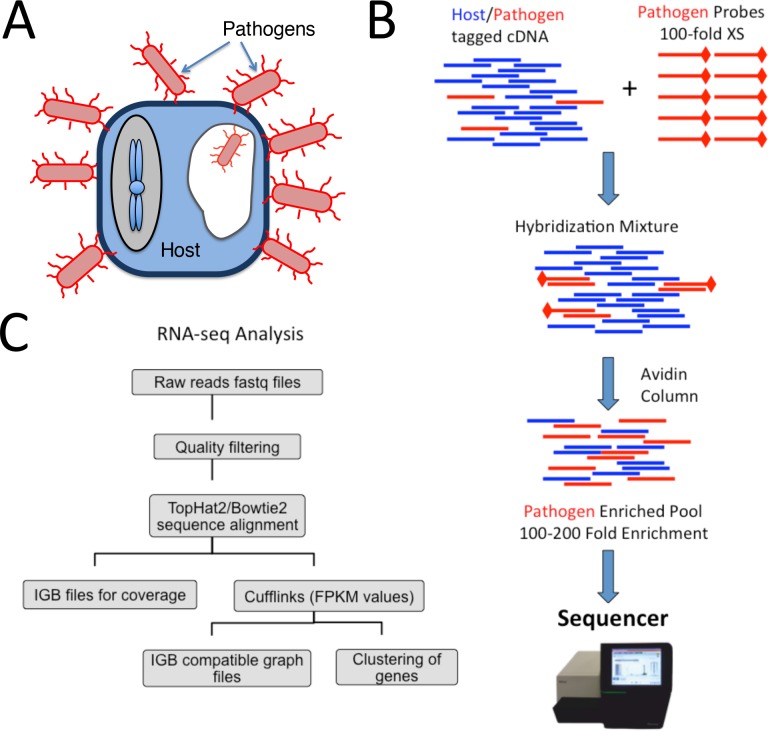
Fig 1. Schematic workflow of capture protocol and analysis.
(A) Infection of pathogen (K. pneumoniae, red) to host P388D1 murine macrophage cells (blue). Non-cell associated bacteria are washed away prior to RNA extraction, such that the bacteria are primarily associated with host cells (both externally and internalized). (B) Capture protocol workflow where host pathogen cDNA with Illumina-compatible adapters is mixed with pathogen-specific capture probes for overnight hybridization. The hybridized mixture is then passed through a biotinylated avidin column to enrich for pathogen transcripts, prior to Illumina library construction. (C) RNA-seq workflow of YAnTra which was used to analyze RNA seq raw data where: Raw reads were quality filtered, aligned to reference genome, and processed with cufflinks to obtain gene expression.