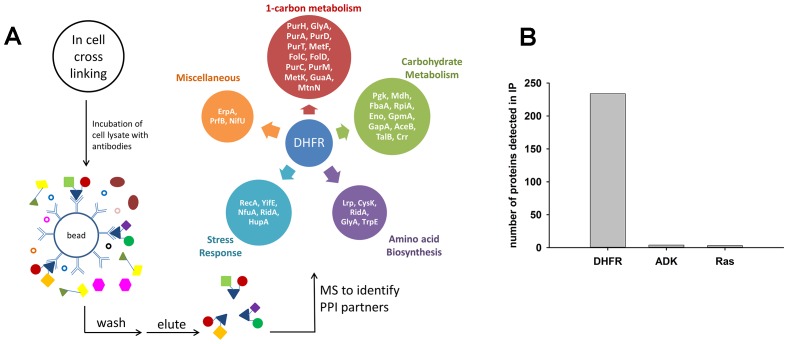
Figure 5. Detection of protein-protein interaction (PPI) partners of E.coli DHFR.
(A) Schematics of the co-immunoprecipitation protocol. Proteins were cross-linked inside the cell using a cell-permeable cross-linker DSP (dithiobis(succinimidyl propionate)), and following lysis, the lysate was captured on anti-DHFR/anti-ADK/anti-Ras/anti-His-tag antibodies to fish out the protein of interest and its PPI partners. The interacting partners were subsequently identified using LC-MS/MS analysis. The observed interactome of overexpressed DHFR could be classified broadly into carbohydrate metabolism and 1-carbon metabolism (purine biosynthesis and amino acid metabolism groups). (B) Over-expressed E. coli DHFR picked out 234 proteins in total, while ADK and Ras detected only 4 and 3 proteins respectively. In case of DHFR, IP datasets were background subtracted using lysate from untransformed WT E. coli cells and DHFR3 (a MG1655 strain where the chromosomal DHFR has been replaced with DHFR from an orthologous bacteria (Bershtein et al., 2015) (also see SI Materials and methods for data analysis). In case of IP for ADK and Ras over-expression, background correction was done with lysate with only untransformed WT E. coli cells. Three biological repeat IP experiments with over-expressed E. coli DHFR and polyclonal anti-DHFR antibody (Figure 5—source data 1) had statistically significant correlation among themselves (average r = 0.8, p<0.001).
DOI: http://dx.doi.org/10.7554/eLife.20309.013