Figure 1.
Purification of FOXA1-Associated Proteins Using RIME and Mapping of MLL3 Binding Genome-wide
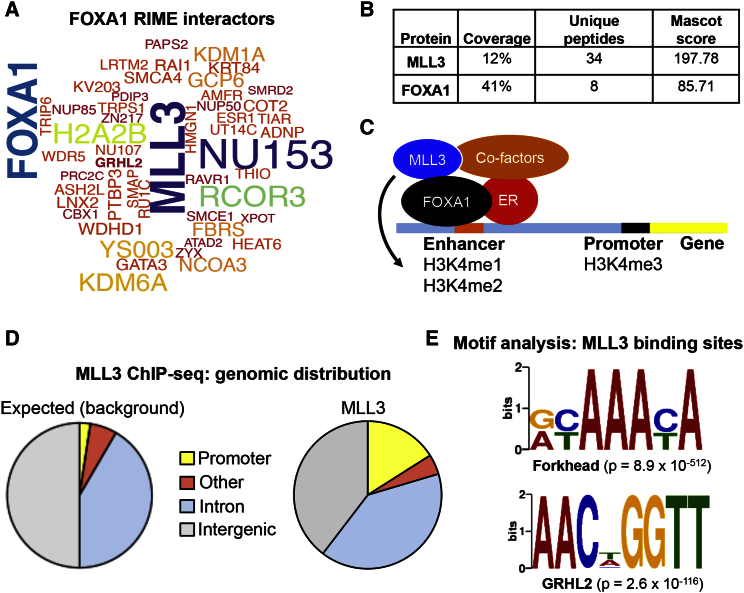
(A) The FOXA1 interactome was discovered by performing RIME in MCF-7 breast cancer cells. The data are represented as a Wordcloud, where the size of protein names represent the strength and confidence of the interactions based on the Mascot score. MLL3 was identified as one of the strongest and most reproducible FOXA1-interacting proteins.
(B) Peptide coverage, number of unique peptides identified, and Mascot score of MLL3 and FOXA1 following FOXA1 purification.
(C) Hypothesized mechanism of FOXA1 and MLL3 function. Our finding that MLL3 and FOXA1 physically interact in breast cancer cells implies that FOXA1 could recruit the enzyme that can add methyl groups to histone 3 lysine 4. FOXA1-bound enhancers are demarcated by H3K4me1 and H3K4me2.
(D) MLL3 ChIP-seq was conducted and the genomic distribution of MLL3 peaks is shown relative to the whole genome (the expected control values). Regions bound by MLL3 occurred mostly at enhancers rather than promoters.
(E) De novo motif analysis of MLL3 binding sites. Motif analysis revealed an enrichment in Forkhead motif, the canonical motif bound by FOXA1, and motifs for the transcription factor grainyhead-like 2 protein (GRHL2).