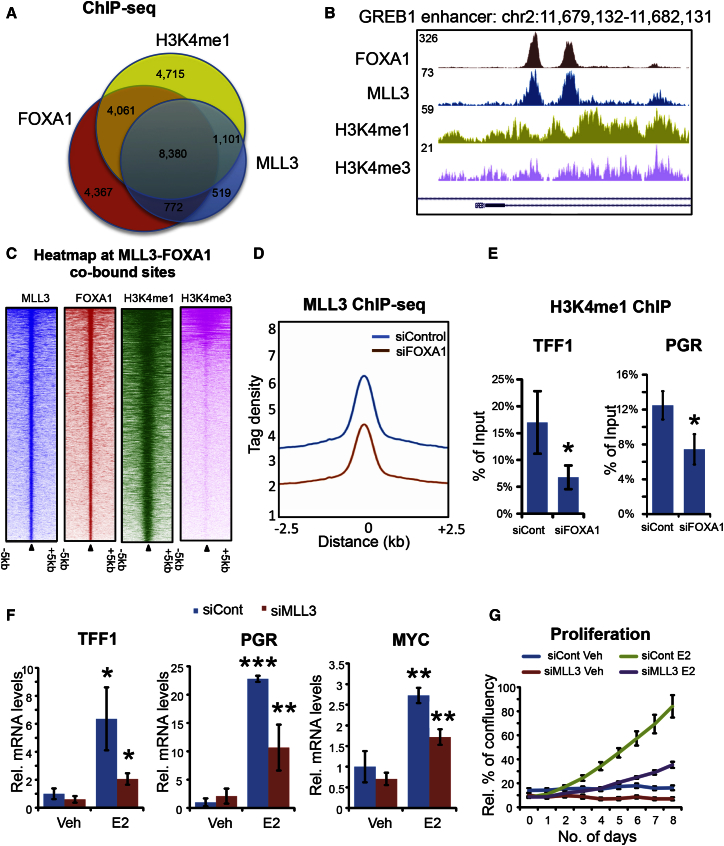
Figure 2.
Co-binding of MLL3, FOXA1, and H3K4me1/me3 and Mechanism of MLL3 Recruitment
(A) Overlap of MLL3, FOXA1, and H3K4me1 binding revealed by ChIP-seq. MLL3 binding sites were co-bound by FOXA1 and the histone marks. The numbers of peaks within each category are shown on the diagram.
(B) An example of an MLL3, FOXA1, and H3K4me1/me3 co-bound region at the GREB1 enhancer.
(C) Heatmap of MLL3-FOXA1 co-bound regions showing binding signal intensity for FOXA1, MLL3, H3K4me1, and H3K4me3. Binding is ranked from the strongest to the weakest binding sites.
(D) Signal intensity plot representing changes in MLL3 ChIP-seq signal in siControl versus siFOXA1-transfected conditions. Differentially bound sites needed to be detected in at least two replicates to be included.
(E) ChIP-qPCR analyses of H3K4me1 after knockdown of FOXA1 on ER-bound enhancers of TFF1 and PGR. n = 3; mean ± SD is shown as the of percentage of input. ∗p ≤ 0.05.
(F) qRT-PCR of estrogen-induced genes TFF1 and PGR with or without knockdown of MLL3 after 3 days of charcoal-stripped serum ±10 nM estrogen (E2) treatment. n = 3; mean ± SD is shown in average relative mRNA levels compared to the vehicle (Veh) condition. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(G) Estrogen-induced proliferation assays with or without knockdown of MLL3 after 3 days of charcoal-stripped serum ±10 nM estrogen treatment for 8 days. n = 4; mean ± SEM of percentage of confluency is shown.