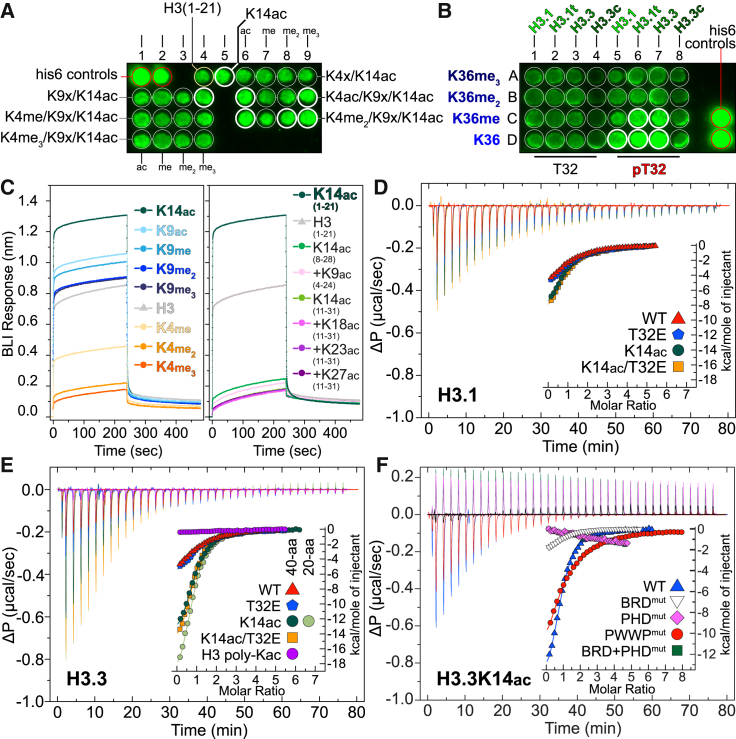
Figure 2.
ZMYND8 Binds to Histone H3 In Vitro
(A) Focused peptide SPOT array spanning lysine acetylation/methylation combinations within the first 21 residues of N-terminal histone H3 peptides. The presence of K14ac increases binding intensity (thick white circles).
(B) Focused peptide SPOT array spanning residues 22–42 of all histone H3.x variants, carrying different methyl states of K36, in the absence or presence of pT32.
(C) BLI of the recombinant triple reader ensemble profiled against 20-aa-long N-terminal histone H3 peptides carrying single PTMs as indicated in the inset (left). Unmodified H3 peptides bind to the triple reader modules (shown in gray). K9 modifications are tolerated, whereas K4 methylations break the interaction, and K14 acetylation greatly enhances it. Removal of the N-terminal sequence from peptides results in loss of binding irrespective of additional modifications (K9ac, K18ac, K23ac, and K27ac) present together with the central K14ac mark (right), suggesting that the N-terminal portion of H3, which engages the PHD domain, is essential for binding to ZMYND8. Experiments were carried out twice (n = 2) for each condition/peptide.
(D) In-solution evaluation of histone H3.1 binding by ITC. Raw injection heats for titrations of modified peptides (carrying specific modifications as indicated in the inset) into a solution of ZMYND8 are shown. The inset shows the normalized binding enthalpies corrected for the heat of peptide dilution as a function of binding site saturation (symbols as indicated in the figure). Solid lines represent a nonlinear least-squares fit using a single-site binding model. Histone H3.1 lacking any modifications binds weakly (red triangle, KD = 19.0 μM), whereas the addition of the phosphomimetic T32E has little effect (blue pentagon, KD = 20.7 μM). Binding to K14ac is also weak (dark green circles, KD = 15.1 μM), and the co-existence of the phosphomimetic T32E shows little effect (orange square, KD = 17.5 μM). ITC titrations were carried out in triplicate (n = 3), and representative curves are shown.
(E) In solution evaluation of histone H3.3. Data are presented as in (D). Histone H3.3 peptides lacking any modifications bind weakly (red triangle, KD = 19.6 μM), whereas the addition of the phosphomimetic T32E has little effect on binding (blue pentagon, KD = 17.4 μM). Binding is mainly driven by K14ac (dark and light green circles, KD = 6.8 μM or 6.6 μM 40-mer and 20-mer, respectively), and the co-existence of the phosphomimetic T32E shows little effect (orange square, KD = 6.2 μM).
(F) In-solution evaluation of ZMYND8 carrying specific domain mutations to histone H3.3 binding by ITC. Data are presented as in (D). Mutation of the conserved asparagine (N228) on the bromodomain abolishes the interaction, as do mutations of conserved PHD residues (N87A, E104A, and D124A). Combination of PHD/BRD mutations has the same effect. Mutation of the conserved residues forming the PWWP cage (F288A and W291A) result in loss of affinity (WT ZMYND8, 6.8 μM; PWWP mutant, 23 μM).