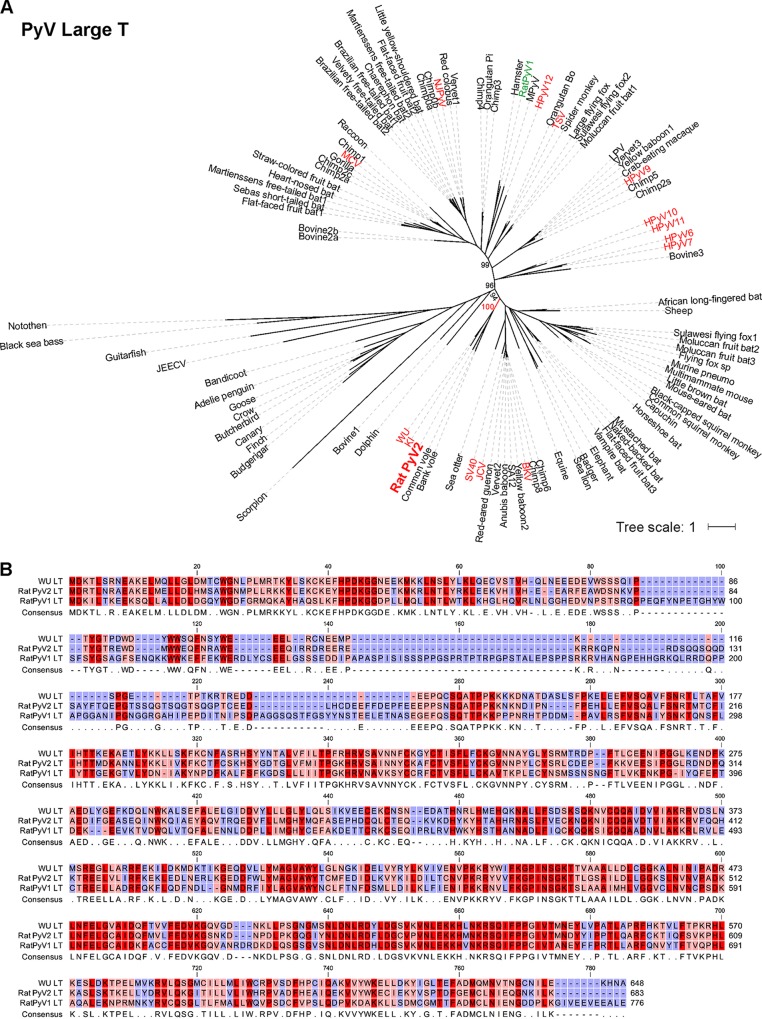
FIG 5 .
Phylogenetic analysis of 103 full-length PyV LT protein sequences. (A) Consensus tree of 1,000 bootstrap replicates was generated by the neighbor-joining method using MEGA7. Percent bootstrap values are given for selected nodes. All known P-PIT-reactive PyV T antigens are shown in red. RatPyV1 is shown in green. The phylogenetic tree was visualized using iTOL. (B) LT amino acid sequence alignment of WU, RatPyV1, and RatPyV2 sequences was generated using CLC Genomics Workbench software. Residue conservation ranges from red (conserved) to pink (mostly conserved) to blue (divergent). A minus sign indicates missing residue at the given position.