Abstract
Mediator is a highly conserved protein complex that functions as a transcriptional coactivator in RNA polymerase II (RNAPII)-mediated transcription. The Arabidopsis Mediator complex has recently been implicated in plant immune responses. Here, we compared salicylic acid (SA)-, methyl jasmonate (MeJA)-, and the ethylene (ET) precursor 1-aminocyclopropane-1-carboxylic acid (ACC)-induced defense and/or wound-responsive gene expression in 14 Arabidopsis Mediator subunit mutants. Our results show that MED14, MED15, and MED16 are required for SA-activated expression of the defense marker gene PATHOEGNESIS-RELATED GENE1, MED25 is required for MeJA-induced expression of the wound-responsive marker gene VEGATATIVE STORAGE PROTEIN1 (VSP1), MED8, MED14, MED15, MED16, MED18, MED20a, MED25, MED31, and MED33A/B (MED33a and MED33B) are required for MeJA-induced expression of the defense maker gene PLANT DEFENSIN1.2 (PDF1.2), and MED8, MED14, MED15, MED16, MED25, and MED33A/B are also required for ACC-triggered expression of PDF1.2. Furthermore, we investigated the involvement of MED14, MED15, and MED16 in plant defense signaling crosstalk and found that MED14, MED15, and MED16 are required for SA- and ET-mediated suppression of MeJA-induced VSP1 expression. This result suggests that MED14, MED15, and MED16 not only relay defense signaling from the SA and JA/ET defense pathways to the RNAPII transcription machinery, but also fine-tune defense signaling crosstalk. Finally, we show that MED33A/B contributes to the necrotrophic fungal pathogen Botrytis cinerea-induced expression of the defense genes PDF1.2, HEVEIN-LIKE, and BASIC CHITINASE and is required for full-scale basal resistance to B. cinerea, demonstrating a positive role for MED33 in plant immunity against necrotrophic fungal pathogens.
Keywords: MED14, MED15, MED16, salicylic acid, jasmonate, and ethylene, defense signaling crosstalk, Arabidopsis
Background
Salicylic acid (SA), jasmonates (JAs), and ethylene (ET) are the primary defense signal molecules of the plant immune system (Pieterse et al., 2009). SA activates resistance against biotrophs and hemibiotrophs, JA mediates defense against necrotrophs and responses to wounding and herbivores, and ET contributes to defense signaling against necrotrophs (Pozo et al., 2004; van Loon et al., 2006; Loake and Grant, 2007). While each of these signal molecules induces a specific defense signaling pathway(s), there is extensive crosstalk among them (Thomma et al., 2001; Glazebrook, 2005; Pieterse et al., 2009). For instance, SA and JA signaling mostly antagonize each other, and ET enhances both SA- and JA-mediated defense responses against pathogens, but suppresses JA-mediated wound signaling. Such crosstalk allows plants to prioritize one defense response over others when encountering a specific attacker. Defense signaling crosstalk has been extensively studied in recent years (Pieterse et al., 2009), but the underlying molecular mechanisms still await full characterization.
Mediator is a highly conserved protein complex that is essential for RNA polymerase II (RNAPII)-mediated transcription (Conaway and Conaway, 2011). This protein complex exists in multiple functionally distinct forms and acts as either a transcriptional activator or a repressor, depending on its associated protein partners. The Mediator core contains more than 20 subunits, which are organized into head, middle, and tail modules (Guglielmi et al., 2004; Chadick and Asturias, 2005). Mediator associates with the RNAPII complex via the head and middle modules to form the holoenzyme, which stimulates basal transcription and supports activation of transcription by specific transcriptional activators (Ansari et al., 2009). By interacting with particular transcriptional activators, individual Mediator subunits converge diverse signals to the RNAPII transcription complex, leading to pathway-specific gene transcription (Balamotis et al., 2009). The head and middle modules of Mediator can also interact with a kinase module, which prevents their binding to the RNAPII complex, leading to transcriptional repression (Knuesel et al., 2009). The Arabidopsis Mediator complex contains 27 conserved subunits and six additional subunits whose positions in the complex are unassigned (Bäckström et al., 2007; Mathur et al., 2011). A number of the Arabidopsis Mediator subunits have been implicated in immune responses. For instance, MED14, MED15, MED16, and MED19a have been shown to regulate the SA-triggered immunity against biotrophic and hemibiotrophic pathogens (Canet et al., 2012; Zhang et al., 2012, 2013; Caillaud et al., 2013), whereas MED8, MED12, MED13, MED14, MED16, MED21, MED25, and CDK8 have been found to function in JA/ET-mediated immunity against necrotrophic pathogens (Dhawan et al., 2009; Kidd et al., 2009; Zhang et al., 2012; Zhu et al., 2014). MED18 also functions in resistance to necrotrophic pathogens, but the resistance appears to be independent of the JA/ET signaling (Lai et al., 2014).
We have previously shown that the Arabidopsis Mediator complex subunit MED16 is required for ET-promoted inhibition of JA-mediated wound signaling (Wang et al., 2015b), indicating that some of the Mediator subunits may be involved in defense signaling crosstalk. Here, we compared SA-, methyl jasmonate (MeJA)-, and the ET precursor 1-aminocyclopropane-1-carboxylic acid (ACC)-induced defense and/or wound-responsive marker gene expression in 14 Arabidopsis Mediator subunit mutants and identified MED14, MED15, and MED16 as key players in plant defense signaling crosstalk. Additionally, we found that the Mediator subunits MED33A and MED33B (MED33A/B) positively contribute to Arabidopsis defense responses against the necrotrophic fungal pathogen Botrytis cinerea.
Materials and Methods
Plant Materials and Growth Conditions
The wild-type used in this study was the Arabidopsis thaliana (L.) Heynh. ecotype Columbia (Col-0). All Mediator mutants except med33b were previously described (Wang et al., 2015b). The med33b mutant (SALK_037472) and med15/nbr4-4 (SAIL_792_F02) were obtained from the Arabidopsis Biological Resource Center at The Ohio State University (Columbus, OH, USA). Homozygous mutant plants of SALK_037472 were confirmed with primers (forward: 5′GTACGAGGTTGCAACTACTG3′and reverse: 5′GCAGTGGAGAAAACAGCATG3′) that flank the T-DNA insertion and the left border primer LBa1 (Alonso et al., 2003). The med33a/b double mutant was created by crossing SALK_037472 with SALK_022477 (med33a) and identified in the F2 generation by PCR. The Arabidopsis seeds were sown on autoclaved soil (Sunshine MVP; Sun Gro Horticulture, Agawam, MA, USA) and cold-treated at 4°C for 3 days. Plants were germinated and grown at 22–24°C under a 16-hr-light/8-hr-dark regime. Four-week-old soil-grown plants were used for pathogen infection.
Chemical Treatment
Ten-day-old seedlings grown on one-half-strength Murashige and Skoog (½12 × MS) medium were treated with 0.5 mM SA, 0.1 mM MeJA, 0.1 mM ACC, or their combination. Seedlings for the negative control were treated with water. Aerial parts of the seedlings were collected and subjected to total RNA extraction.
Pathogen Infection
The B. cinerea strain B05 was used in this study. B. cinerea inoculation and lesion size measurement were conducted as described in detail previously (Wang et al., 2015a).
RNA Analysis
Total RNA extraction, reverse transcription, and real-time quantitative PCR (qPCR) were performed as previously described (Defraia et al., 2010). Primers used for PR1, VSP1, and PDF1.2 were described previously (Defraia et al., 2010; Wang et al., 2015b). Primers used for HEL are forward: 5′GTGAGTGCTTATTGCTCCAC3′ and reverse: 5′ACATCCAAATCCAAGCCTCC3′, and for CHIB are forward: 5′GGTTCTGGATGACTGCTCAG3′ and reverse: 5′CTATACGATCGGCGACTCTC3′.
Statistical Methods
Statistical analyses were performed using the one-way ANOVA and the two-way ANOVA in Prism 5.0b (GraphPad Software, La Jolla, CA, USA). Lesion sizes measured in three independent experiments were combined and analyzed as a one-way ANOVA, blocked by experiment, using JMP 11 (JMP Software, Cary, NC, USA). All experiments were repeated three independent times with similar trends. Results from a representative experiment are presented.
Results
SA-, MeJA, and ACC-Induced Defense Marker Gene Expression in 14 Mediator Mutants
To compare the function of different Arabidopsis Mediator subunits in the SA, JA, and ET signaling pathways, we tested SA-induced expression of the SA pathway marker gene PATHOEGNESIS-RELATED GENE1 (PR1), MeJA-induced expression of the wound-responsive marker gene VEGATATIVE STORAGE PROTEIN1 (VSP1) and the defense marker gene PLANT DEFENSIN1.2 (PDF1.2), and ACC-induced expression of PDF1.2 in the previously described 13 Mediator subunit mutants except that a med33a/b double mutant was used to replace the med33b single mutant (Wang et al., 2015b). Ten-day-old seedlings of the wild-type Col-0 and the 13 Mediator mutants grown on ½12 × MS medium were treated with SA, MeJA, or ACC. We also included the med15/nrb4-4 mutant in the experiment, as MED15 is essential for SA signaling (Canet et al., 2012). Since homozygous med15 plants are sterile, we used seeds from heterozygous plants. Three weeks after germination in soil, the small and chlorotic homozygous med15 plants were transplanted and allowed to grow for four more weeks. As med15 mutant plants grow very slowly compared with Col-0, 3-week-old soil-grown Col-0 plants with a size similar to that of the med15 plants were used for comparison. As shown Figure 1A, SA-induced PR1 expression was significantly blocked in med14, med15, and med16, MeJA-induced VSP1 expression was significantly reduced only in med25, MeJA-induced PDF1.2 expression was significantly decreased in med8, med14, med15, med16, med18, med20a, med25, med31, and med33a/b, and ACC-induced PDF1.2 expression was significantly inhibited in med8, med14, med15, med16, med25, and med33a/b. Note that the observation that MeJA-induced PDF1.2 expression was significantly decreased in med18 is in contrast to the previous report (Lai et al., 2014). This discrepancy is probably due to different growth conditions. Nevertheless, these results indicate that, among the 14 Mediator subunits, MED14, MED15, and MED16 are required for SA-activated PR1 expression, MED25 is required for MeJA-induced VSP1 expression, MED8, MED14, MED15, MED16, MED18, MED20a, MED25, MED31, and MED33A/B are required for MeJA-induced PDF1.2 expression, and MED8, MED14, MED15, MED16, MED25, and MED33A/B are required for ACC-induced PDF1.2 expression.
FIGURE 1.
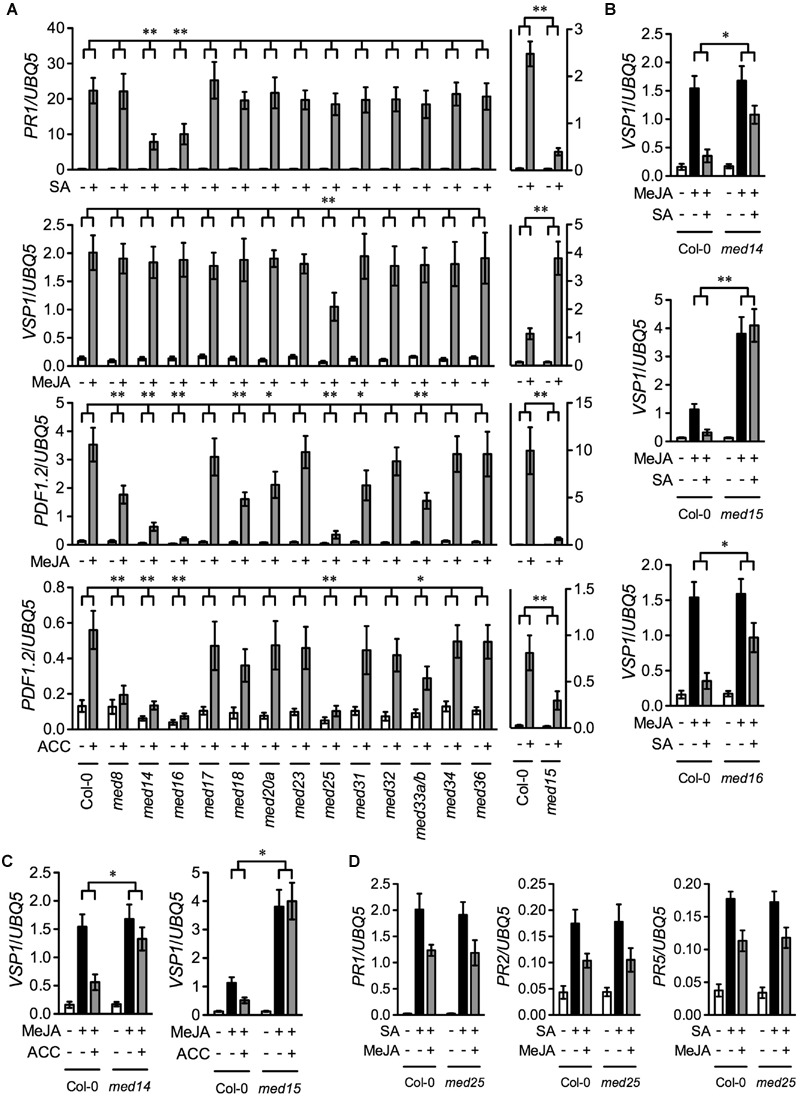
SA-, MeJA-, ACC-, and their combinations-induced pathogen- and wound-responsive genes in Mediator subunit mutants. (A) SA-induced PR1, MeJA-induced VSP1 and PDF1.2, and ACC-induced PDF1.2 in 14 Mediator subunit mutants. Ten-day-old seedlings of Col-0 and the indicated Mediator mutants except med15 grown on ½12 × MS medium as well as 3-week-old soil-grown Col-0 and 7-week-old soil-grown med15 plants were treated with 0.5 mM SA, 0.1 mM MeJA, or 0.1 mM ACC. Plant tissues were collected 6 h after the treatment for analysis of VSP1 and 24 h for PR1 and PDF1.2. (B) SA-mediated inhibition of MeJA-induced expression of VSP1 in med14, med15, and med16. Ten-day-old Col-0, med14, and med16 seedlings grown on ½12 × MS medium as well as 3-week-old soil-grown Col-0 and 7-week-old soil-grown med15 plants were treated with 0.1 mM MeJA or 0.1 mM MeJA plus 0.5 mM SA. Plant tissues were collected 6 h after the treatment. (C) ET-mediated inhibition of MeJA-induced expression of VSP1 in med14 and med15. Ten-day-old Col-0 and med14 seedlings grown on ½12 × MS medium as well as 3-week-old soil-grown Col-0 and 7-week-old soil-grown med15 plants were treated with 0.1 mM MeJA or 0.1 mM MeJA plus 0.1 mM ACC. Plant tissues were collected 6 h after the treatment. (D) SA-induced PR gene expression in med25 in the presence and absence of MeJA. Ten-day-old Col-0 and med25 seedlings grown on ½12 × MS medium were treated with 0.5 mM SA or 0.5 mM SA plus 0.1 mM MeJA. Plant tissues were collected 24 h after the treatment. Total RNA was extracted from the collected plant tissues and subjected to real-time qPCR analysis. Expression of the target genes was normalized against the constitutively expressed UBQ5. Data represent means of three biological replicates with standard deviation (SD). Asterisks indicate that the induction of the gene was significantly lower or higher (A) and the inhibition of MeJA-induced VSP1 expression was significantly weaker (B,C) in the mutant than in the Col-0 plants (∗P < 0.05, ∗∗P < 0.01, two-way ANOVA).
Involvement of MED14, MED15, and MED16 in Defense Signaling Crosstalk
Since MED14, MED15, and MED16 function in the SA pathway (Canet et al., 2012; Wathugala et al., 2012; Zhang et al., 2012, 2013), they might also be involved in SA-mediated suppression of JA signaling. To test this hypothesis, we treated med14, med15, med16, and Col-0 plants with MeJA or MeJA plus SA and examined the induction of the MeJA-induced wound-responsive gene VSP1. As shown in Figure 1B, SA significantly inhibited MeJA-induced expression of VSP1 in the Col-0 plants, but the inhibition was significantly alleviated in med14 and med16, and completely blocked in med15, indicating that MED14, MED15, and MED16 are all required for SA-mediated suppression of JA-mediated wound-responsive gene expression. Moreover, MED14, MED15, and MED16 also function in the ET-mediated defense pathway. As MED16 is required for ET-activated suppression of JA-mediated wound signaling (Wang et al., 2015b), MED14 and MED15 might also be required for this process. To test this, we treated med14, med15, and Col-0 plants with MeJA or MeJA plus ACC and tested the induction of VSP1. As shown in Figure 1C, ACC significantly inhibited MeJA-induced expression of VSP1 in the Col-0 plants, but the inhibition was dramatically relieved in med14 and completely blocked in med15. Therefore, as MED16 (Wang et al., 2015b), MED14 and MED15 are also required for ET-mediated suppression of JA-induced wound-responsive gene expression. Finally, since MED25 functions in JA-mediated pathogen and wound responses (Kidd et al., 2009; Chen et al., 2012), it might modulate JA-mediated suppression of SA signaling. To test this hypothesis, we treated med25 and Col-0 plants with SA and SA plus MeJA and examined the induction of the SA-responsive genes PR1, PR2, and PR5. As shown in Figure 1D, MeJA inhibited SA-induced PR gene expression to similar extents in the Col-0 and med25 plants, indicating that MED25 is not involved in JA-mediated suppression of SA signaling.
Function of MED33A/B in Basal Resistance against the Necrotrophic Fungal Pathogen B. cinerea
PDF1.2 is a marker gene of the JA/ET-mediated defense signaling, which is central in resistance to necrotrophic pathogens. The Mediator subunits MED8, MED14, MED16, MED18, and MED25 are required for MeJA- and/or ACC-induced PDF1.2 expression and contribute to resistance to necrotrophic fungal pathogens (Kidd et al., 2009; Wathugala et al., 2012; Zhang et al., 2012; Lai et al., 2014; Wang et al., 2015b). Since MED20a, MED31, and MED33A/B are also required for full induction of PDF1.2 by MeJA and/or ACC, we examined B. cinerea-induced expression of three JA/ET-responsive genes PDF1.2, HEVEIN-LIKE (HEL), and BASIC CHITINASE (CHIB) in med20a, med31, and med33a/b and tested resistance of these mutants to B. cinerea. We did not include the med15 mutant in the experiment due to its extremely delayed growth. As shown in Figure 2A, B. cinerea-induced expression of PDF1.2, HEL, and CHIB was significantly reduced only in the med33a/b double mutant. Consistently, med33a/b also exhibited enhanced susceptible to B. cinerea (Figures 2B,C). These results indicate that MED33A/B plays a positive role in defense against this necrotrophic fungal pathogen.
FIGURE 2.
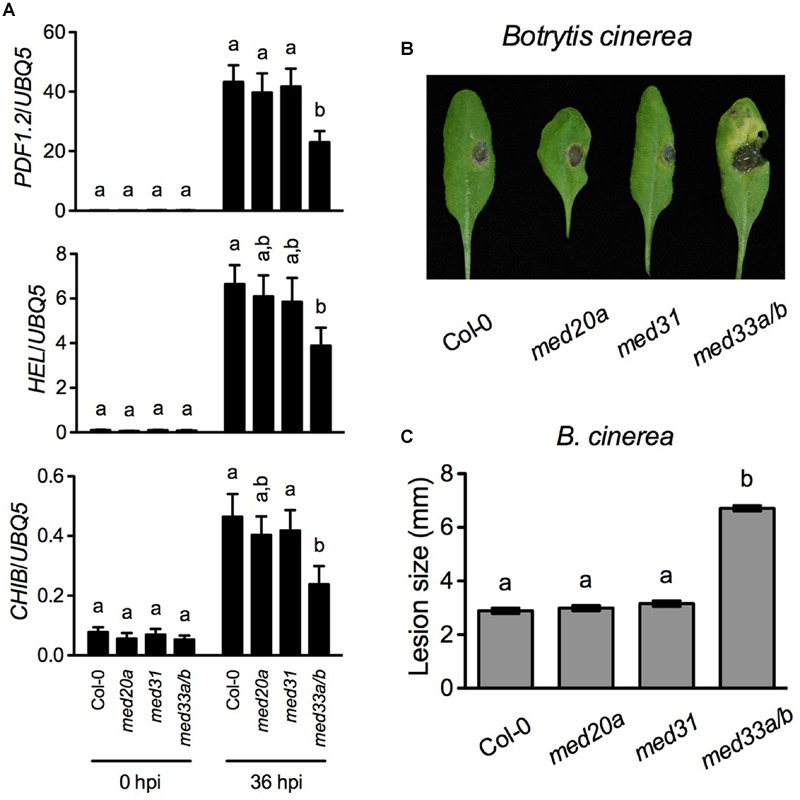
Botrytis cinerea-induced defense responses in med20a, med31, and med33a/b. (A) B. cinerea-induced expression of PDF.2, HEL, and CHIB in Col-0, med20a, med31, and med33a/b plants. Leaf tissues were collected 36 h post-inoculation (hpi). RNA extraction and real-time qPCR were performed as in Figure 1. Data represent means of three biological replicates with SD. Different letters above the bars indicate significant differences (P < 0.05, on-way ANOVA). The statistical comparisons were performed among genotypes for each time point. (B) Disease symptoms on rosette leaves of 4-week-old soil-grown plants inoculated with B. cinerea. Photos were taken 4 days post-inoculation. (C) Size of the necrotrophic lesions formed on B. cinerea-infected Col-0, med20a, med31, and med33a/b plants. Lesion sizes on 90 leaves measured in three independent experiments were combined and analyzed as a one-way ANOVA, blocked by experiment. The resulting mean and standard error are presented. Different letters above the bars indicate significant differences (P < 0.0001).
Discussion
It is generally accepted that the tail module of Mediator is the main target for transcriptional activators. The Arabidopsis Mediator tail module consists of MED14, MED15, MED16, MED23, MED27, MED32, and MED33 (Mathur et al., 2011). In this study, we characterized T-DNA insertion mutants of all tail module subunits except MED27, for which no homozygous T-DNA insertion line was identified. Our results show that, besides MED16 (Wathugala et al., 2012; Zhang et al., 2012), MED14, MED15, and MED33A/B are also required for full induction of the defense marker gene PDF1.2 by MeJA and ACC (Figure 1A). MED33A/B also contributes to B. cinerea-induced defense gene expression and is required for full-scale basal resistance against this necrotrophic fungal pathogen (Figures 2A–C).
Importantly, we found that the tail module subunits MED14, MED15, and MED16 not only play dominant roles in regulation of both SA- and JA/ET-mediated defense responses (Canet et al., 2012; Wathugala et al., 2012; Zhang et al., 2012, 2013), but also are required for both SA- and ET-promoted inhibition of JA-mediated wound signaling (Figures 1B,C). These results indicate that MED14, MED15, and MED16 not only relay defense signaling from the SA and JA/ET pathways to the RNAPII transcription machinery, but also fine-tune defense-related transcriptional changes. We have recently shown that the transcription factor WRKY33, which is an important regulator of defense against necrotrophic fungal pathogens (Zheng et al., 2006), delivers signals to Mediator by interacting with MED16 (Wang et al., 2015b). The SA pathway transcriptional coactivator NONEXPRESSER OF PR GENE1 and TGA transcription factors as well as the JA/ET defense pathway transcription factors ETHYLENE INSENSITIVE3 (EIN3), ETHYLENE INSENSITIVE3-LIK1 (EIL1), OCTADECANOID-RESPONSIVE ARABIDOPSIS AP2/ERF59 (ORA59), and ETHYLENE RESPONSIVE FACTOR (ERF1) may also deliver signals to Mediator through the tail module. Indeed, it has been shown that EIN3, EIL1, ORA59, and ERF1 all interact with MED25, which in turn is physically associated with MED16 (Cevik et al., 2012; Yang et al., 2014). Whether any of the SA and JA/ET defense pathway transcriptional activators interact directly with MED14, MED15, and/or MED16 awaits further investigation.
Author Contributions
ZM and CW conceived and designed the experiments. CW and XD performed the experiments. CW and ZM analyzed the data. ZM and CW wrote the paper. All of the authors carefully checked an approved this version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Arabidopsis Biological Resource Center at The Ohio State University (Columbus, OH, USA) for SALK_006603, SALK_111977, SAIL_889_C08, SALK_119080, SALK_035522, SALK_028490, SALK_022477, SALK_087178, and SALK_093373 seeds, and the European Arabidopsis Stock Centre at The University of Nottingham (Nottingham, UK) for GABI_507F08 seeds.
Footnotes
Funding. This work was supported by the National Sclerotinia Initiative (grant no. 58-5442-3-029).
References
- Alonso J. M., Stepanova A. N., Leisse T. J., Kim C. J., Chen H., Shinn P., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. 10.1126/science.1086391 [DOI] [PubMed] [Google Scholar]
- Ansari S. A., He Q., Morse R. H. (2009). Mediator complex association with constitutively transcribed genes in yeast. Proc. Natl. Acad. Sci. U.S.A 106 16734–16739. 10.1073/pnas.0905103106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckström S., Elfving N., Nilsson R., Wingsle G., Bjorklund S. (2007). Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol. Cell 26 717–729. 10.1016/j.molcel.2007.05.007 [DOI] [PubMed] [Google Scholar]
- Balamotis M. A., Pennella M. A., Stevens J. L., Wasylyk B., Belmont A. S., Berk A. J. (2009). Complexity in transcription control at the activation domain-mediator interface. Sci. Signal. 2:ra20 10.1126/scisignal.1164302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillaud M. C., Asai S., Rallapalli G., Piquerez S., Fabro G., Jones J. D. (2013). A downy mildew effector attenuates salicylic acid-triggered immunity in Arabidopsis by interacting with the host mediator complex. PLoS Biol. 11:e1001732 10.1371/journal.pbio.1001732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canet J. V., Dobon A., Tornero P. (2012). Non-recognition-of-BTH4, an Arabidopsis mediator subunit homolog, is necessary for development and response to salicylic acid. Plant Cell 24 4220–4235. 10.1105/tpc.112.103028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevik V., Kidd B. N., Zhang P., Hill C., Kiddle S., Denby K. J., et al. (2012). MEDIATOR25 acts as an integrative hub for the regulation of jasmonate-responsive gene expression in Arabidopsis. Plant Physiol. 160 541–555. 10.1104/pp.112.202697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadick J. Z., Asturias F. J. (2005). Structure of eukaryotic mediator complexes. Trends Biochem. Sci. 30 264–271. 10.1016/j.tibs.2005.03.001 [DOI] [PubMed] [Google Scholar]
- Chen R., Jiang H., Li L., Zhai Q., Qi L., Zhou W., et al. (2012). The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 24 2898–2916. 10.1105/tpc.112.098277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway R. C., Conaway J. W. (2011). Function and regulation of the Mediator complex. Curr. Opin. Genet. Dev. 21 225–230. 10.1016/j.gde.2011.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defraia C. T., Zhang X., Mou Z. (2010). Elongator subunit 2 is an accelerator of immune responses in Arabidopsis thaliana. Plant J. 64 511–523. 10.1111/j.1365-313X.2010.04345.x [DOI] [PubMed] [Google Scholar]
- Dhawan R., Luo H., Foerster A. M., Abuqamar S., Du H. N., Briggs S. D., et al. (2009). HISTONE MONOUBIQUITINATION1 interacts with a subunit of the mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell 21 1000–1019. 10.1105/tpc.108.062364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43 205–227. 10.1146/annurev.phyto.43.040204.135923 [DOI] [PubMed] [Google Scholar]
- Guglielmi B., van Berkum N. L., Klapholz B., Bijma T., Boube M., Boschiero C., et al. (2004). A high resolution protein interaction map of the yeast mediator complex. Nucleic Acids Res. 32 5379–5391. 10.1093/nar/gkh878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd B. N., Edgar C. I., Kumar K. K., Aitken E. A., Schenk P. M., Manners J. M., et al. (2009). The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell 21 2237–2252. 10.1105/tpc.109.066910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuesel M. T., Meyer K. D., Bernecky C., Taatjes D. J. (2009). The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes Dev. 23 439–451. 10.1101/gad.1767009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z., Schluttenhofer C. M., Bhide K., Shreve J., Thimmapuram J., Lee S. Y., et al. (2014). MED18 interaction with distinct transcription factors regulates multiple plant functions. Nat. Commun. 5:3064 10.1038/ncomms4064 [DOI] [PubMed] [Google Scholar]
- Loake G., Grant M. (2007). Salicylic acid in plant defence-the players and protagonists. Curr. Opin. Plant Biol. 10 466–472. 10.1016/j.pbi.2007.08.008 [DOI] [PubMed] [Google Scholar]
- Mathur S., Vyas S., Kapoor S., Tyagi A. K. (2011). The Mediator complex in plants: structure, phylogeny, and expression profiling of representative genes in a dicot (Arabidopsis) and a monocot (rice) during reproduction and abiotic stress. Plant Physiol. 157 1609–1627. 10.1104/pp.111.188300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse C. M., Leon-Reyes A., Van der Ent S., Van Wees S. C. (2009). Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5 308–316. 10.1038/nchembio.164 [DOI] [PubMed] [Google Scholar]
- Pozo M. J., Van Loon L. C., Pieterse C. M. J. (2004). Jasmonates-signals in plant-microbe interactions. J. Plant Growth Regul. 23 211–222. 10.1007/BF02637262 [DOI] [Google Scholar]
- Thomma B. P., Penninckx I. A., Broekaert W. F., Cammue B. P. (2001). The complexity of disease signaling in Arabidopsis. Curr. Opin. Immunol. 13 63–68. 10.1016/S0952-7915(00)00183-7 [DOI] [PubMed] [Google Scholar]
- van Loon L. C., Geraats B. P., Linthorst H. J. (2006). Ethylene as a modulator of disease resistance in plants. Trends Plant Sci. 11 184–191. 10.1016/j.tplants.2006.02.005 [DOI] [PubMed] [Google Scholar]
- Wang C., Ding Y., Yao J., Zhang Y., Sun Y., Colee J., et al. (2015a). Arabidopsis elongator subunit 2 positively contributes to resistance to the necrotrophic fungal pathogens Botrytis cinerea and Alternaria brassicicola. Plant J. 83 1019–1033. 10.1111/tpj.12946 [DOI] [PubMed] [Google Scholar]
- Wang C., Yao J., Du X., Zhang Y., Sun Y., Rollins J. A., et al. (2015b). The Arabidopsis mediator complex subunit16 is a key component of basal resistance against the necrotrophic fungal pathogen Sclerotinia sclerotiorum. Plant Physiol. 169 856–872. 10.1104/pp.15.00351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wathugala D. L., Hemsley P. A., Moffat C. S., Cremelie P., Knight M. R., Knight H. (2012). The mediator subunit SFR6/MED16 controls defence gene expression mediated by salicylic acid and jasmonate responsive pathways. New Phytol. 195 217–230. 10.1111/j.1469-8137.2012.04138.x [DOI] [PubMed] [Google Scholar]
- Yang Y., Ou B., Zhang J., Si W., Gu H., Qin G., et al. (2014). The Arabidopsis mediator subunit MED16 regulates iron homeostasis by associating with EIN3/EIL1 through subunit MED25. Plant J. 77 838–851. 10.1111/tpj.12440 [DOI] [PubMed] [Google Scholar]
- Zhang X., Wang C., Zhang Y., Sun Y., Mou Z. (2012). The Arabidopsis mediator complex subunit16 positively regulates salicylate-mediated systemic acquired resistance and jasmonate/ethylene-induced defense pathways. Plant Cell 24 4294–4309. 10.1105/tpc.112.103317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yao J., Zhang Y., Sun Y., Mou Z. (2013). The Arabidopsis mediator complex subunits MED14/SWP and MED16/SFR6/IEN1 differentially regulate defense gene expression in plant immune responses. Plant J. 75 484–497. 10.1111/tpj.12216 [DOI] [PubMed] [Google Scholar]
- Zheng Z., Qamar S. A., Chen Z., Mengiste T. (2006). Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 48 592–605. 10.1111/j.1365-313X.2006.02901.x [DOI] [PubMed] [Google Scholar]
- Zhu Y., Schluttenhoffer C. M., Wang P., Fu F., Thimmapuram J., Zhu J. K., et al. (2014). CYCLIN-DEPENDENT KINASE8 differentially regulates plant immunity to fungal pathogens through kinase-dependent and -independent functions in Arabidopsis. Plant Cell 26 4149–4170. 10.1105/tpc.114.128611 [DOI] [PMC free article] [PubMed] [Google Scholar]


