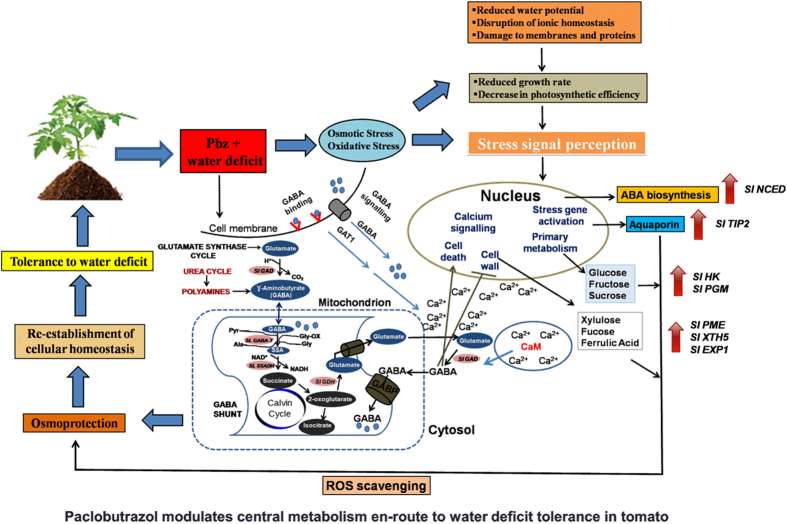
Figure 6. A schematic model describing mechanism of Pbz mediated water deficit tolerance in tomato.
Drought stress in plants is accompanied by secondary oxidative or osmotic stress leading to reduction in water potential, disruption of ionic and osmotic homeostasis and damage to proteins and membranes, ultimately resulting in reduced photosynthetic efficiency and overall growth of the plant. Such a signal, perceived in the nucleus leads to activation of stress responsive genes. Similarly, anti-oxidant genes, genes responsible for ABA biosynthesis as well as genes governing primary metabolism (sugar synthesis, glucose, fructose, sucrose) and constituents of cell wall permeability are induced imparting osmoprotection, re-establishment of cellular homeostasis and tolerance to water deficit. Water deficit stress when applied in conjunction with Pbz engenders modulation of central metabolism through enhanced TCA cycle activity and regulation of gene expression associated with GABA shunt signaling. Generally, external stimuli like drought stress or deficit irrigation described here lead to increase in endogenous GABA levels permitting its adherence to cell-surface binding sites, enabling an interim increase in Ca2+ pools and its import into cells through high affinity GABA transporters (e.g., GAT174). Considerably, GAD is activated through a Ca2+/CaM complex45,75. Thereafter, increased intracellular GABA activates various signaling cascades and genes of primary metabolism (SlHK and SlPGM) while inhibiting some, like genes responsible for cell wall-modifications. Additionally, subject to outside environment, a substantial portion of cytosolic GABA makes way into mitochondria via the GABA permease, At GABP76, for catabolism by GABA-T and SSADH, producing succinate for feeding into the Calvin Cycle. Enzyme represent oval grey boxes, their reactions represented by black arrows. Brown lines indicate a regulatory effect. Blue spheres denote GABA, red crescents denote GABA receptors. Abbreviations: Pbz, Paclobutrazol; GABA, γ-aminobutric acid; GAT1, GABA transporter 1 SlNCED, 9-cis-epoxy-carotenoid dioxygenase; SlTIP2, tonoplast intrinsic protein 2; SlHK, hexokinase; SlPGM, phosphoglucomutase; SlPME, Pectin methyl esterase; SlXTH5, Xyloglucan endotransglycosylase; SlEXP1, expansin1; SlGDH, Glutamate dehydrogenase; CaM, calmodulin; GAD, glutamate decarboxylase; GABA-T, GABA transaminase; SSADH, succinic semialdehyde dehydrogenase; respectively.