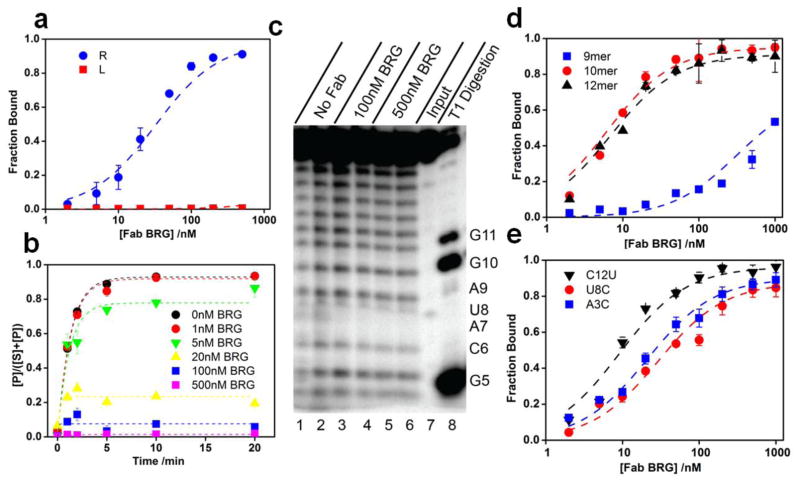
Figure 2.
Location of the Fab-BRG binding site within the branched RNA. (a). Fab BRG binds strand R with full affinity (KD = 26 ± 1 nM). (b). Fab BRG inhibits Dbr1 debranching of the RNA target ([Dbr1] = 1.4 nM). See Supplementary Fig. 1 for original gel figures. (c). Hydroxyl radical footprinting on strand R in the presence and absence of Fab BRG. Input – control samples without Fe-EDTA treatment; T1 Digestion – ladder generated by treating samples with ribonuclease T1. (d). Fab BRG binds a 10-mer oligonucleotide (A3 to C12) with full affinity (KD = 10 ± 2 nM). (e). Fab BRG binds 3 of the 10 scanning mutagenesis 10-mer constructs with nearly full affinity (See Table 1 for more details).