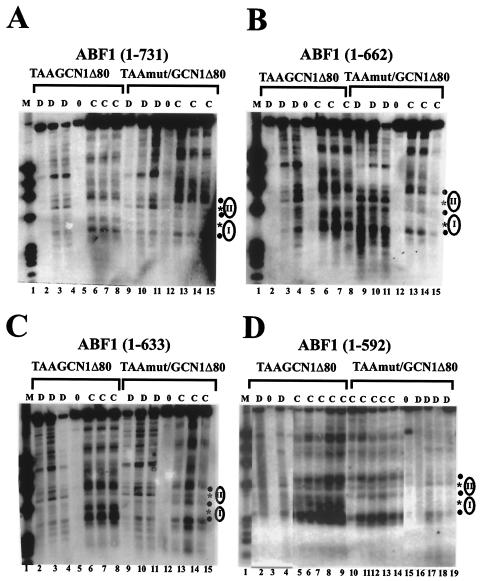
FIG. 3.
Contribution of C-terminal domains of ABF1 to chromatin perturbation via a nucleosomal binding site. (A) MNase cleavage sites in plasmids TAA/GCN1Δ80 and TAAmut/GCN1Δ80 were mapped in GCN4+ yeast harboring ABF1(1-731). Cleavage sites were mapped in naked DNA (D lanes) or in chromatin (C lanes) from the EcoRV site, as in Fig. 1. Lane 1 contains φX/HaeIII marker DNA. Locations of positioned nucleosomes I and II are indicated by ellipses. The filled circles to the right of the panel indicate cleavages enhanced in chromatin relative to DNA, and the stars indicate strong cleavages in naked DNA that are protected by nucleosomes I and II in chromatin of TAAmut/GCN1Δ80 but not TAA/GCN1Δ80. Each lane, beginning with lanes 2 to 4, differs only in the concentration of MNase used. Lanes 5 and 12 show controls not treated with MNase. (B) MNase cleavage sites in plasmids TAA/GCN1Δ80 and TAAmut/GCN1Δ80 were mapped in yeast harboring ABF1(1-662), as in panel A. (C) MNase cleavage sites in plasmids TAA/GCN1Δ80 and TAAmut/GCN1Δ80 were mapped in yeast harboring ABF1(1-633) as in panel A. (D) MNase cleavage sites in plasmids TAA/GCN1Δ80 and TAAmut/GCN1Δ80 were mapped in yeast harboring ABF1(1-592) as in panel A. All samples were run on the same gel, but some lanes are omitted from the figure, as seen from the visible “splice” marks.