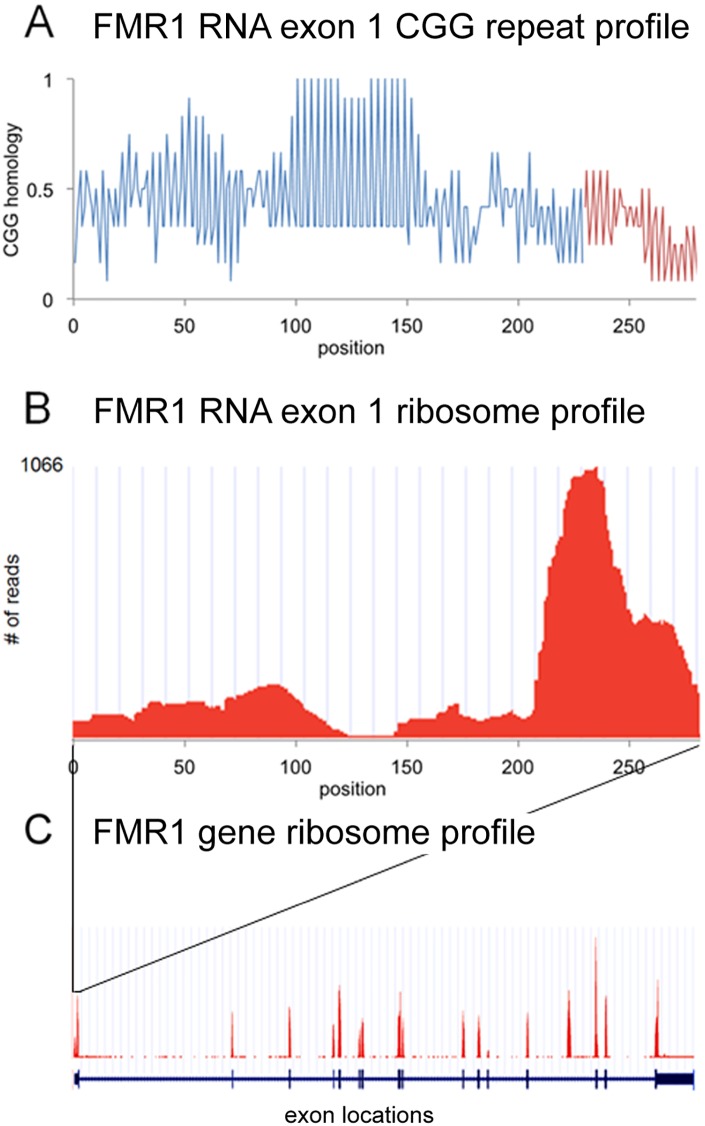
Fig 1. CGG repeat profile and ribosome profile for FMR1 RNA.
Panel A shows the CGG profile for exon 1 of FMR1 RNA, including the 5’UTR (blue) and the initial portion the ORF (red), calculated as described in Materials and Methods. There is a region of CGG repeats between positions 100–150, interrupted by a single AGG at position 131. Panel B shows the corresponding ribosome profile for exon 1 of FMR1 RNA. The number of sequence reads at each position reflects the probability of ribosomes located at that position. Increased ribosome probability downstream of position 200 presumably reflects ribosomes engaged in conventional translation of the FMRP ORF beginning at the AUG at position 230. Increased ribosome probability in upstream regions (1–100 and 150–200) may reflect ribosomes initiating translation at non-canonical AUG-like sites upstream of the CGG repeats or ribosomes engaged in RAN translation of 5’UTR CGG repeats, consistent with reports that translation of CGG repeats in FMR1 RNA may require non-canonical AUG-like sequences upstream of the CGG repeats [11]. The paucity of reads in the region between positions 100–150, which corresponds to the CGG repeat region, may reflect inefficient sequencing through long regions of CGG repeats [24]. Panel C shows the ribosome profile for the entire FMR1 gene (red) and the locations of individual exons (blue), with 5’ and 3’UTR regions indicated by thinner lines and ORF regions indicated by thicker lines. The exon 1 region shown in panels A and B is indicated.