Figure 4. CLIP-170 interactions with formins promote dendritic branching in primary neurons.
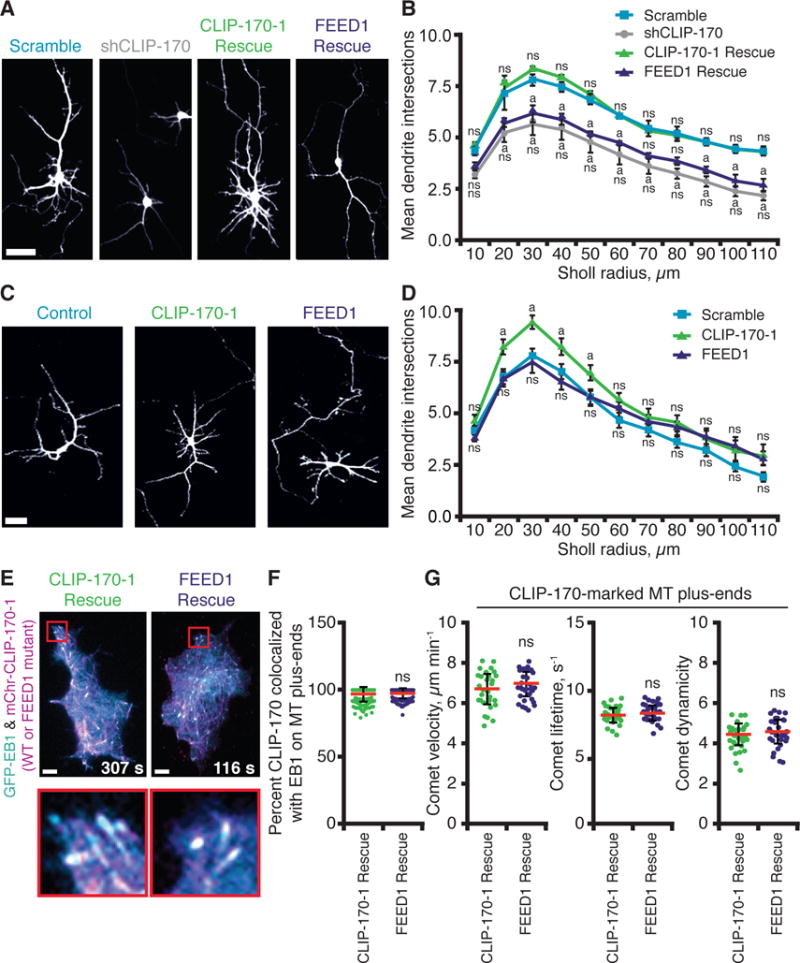
(A) Representative images from 4–5 DIV rat cortical neurons co-transfected with pCMV-GFP and pSUPER containing control (scrambled) shRNA or shRNA against CLIP-170 (shCLIP-170) (16), with or without plasmids expressing shRNA-resistant full-length CLIP-170-1 rescue (wild type or FEED1 mutant). Scale bar, 5 μm. (B) Quantification of dendritic branching complexity by Sholl analysis (n = 90–120 neurons, from two or more independent experiments). Statistical differences: ns, not significantly different from control; a, compared to scramble shRNA control (p < 0.05). There was never a significant difference between the FEED1 rescue the shCLIP-170 knockdown alone. Error bars, SE. (C) Representative images from rat cortical neurons as in (A) expressing full-length CLIP-170-1 (wild type or FEED1 mutant) without silencing endogenous CLIP-170. Scale bar, 5 μm. (D) Quantification of dendritic branching complexity by Sholl analysis as in (B), except data are from n = 60 neurons in two independent experiments. Statistical differences: ns, not significantly different from control; a, compared to scramble shRNA control (p < 0.05). Error bars, SE. (E) Localization of mCherry-CLIP-170-1 (wild type or FEED1 mutant) rescue constructs in N2A cells in which endogenous CLIP-170 was silenced. Scale bars, 10 μm. (F) Quantification of mCherry-CLIP-170-1 colocalization with EB1-GFP at MT plus-ends from cells as in (E). Error bars, SD. (G) Quantitative tracking of mCherry-CLIP-170-1 (wild type or FEED1 mutant) comets on growing MT plus-ends. Analysis includes: comet velocity (or MT growth rate), comet lifetime (MT growth duration), and comet ‘dynamicity’ (a general readout of MT dynamics; (36)). Error bars, SD.
