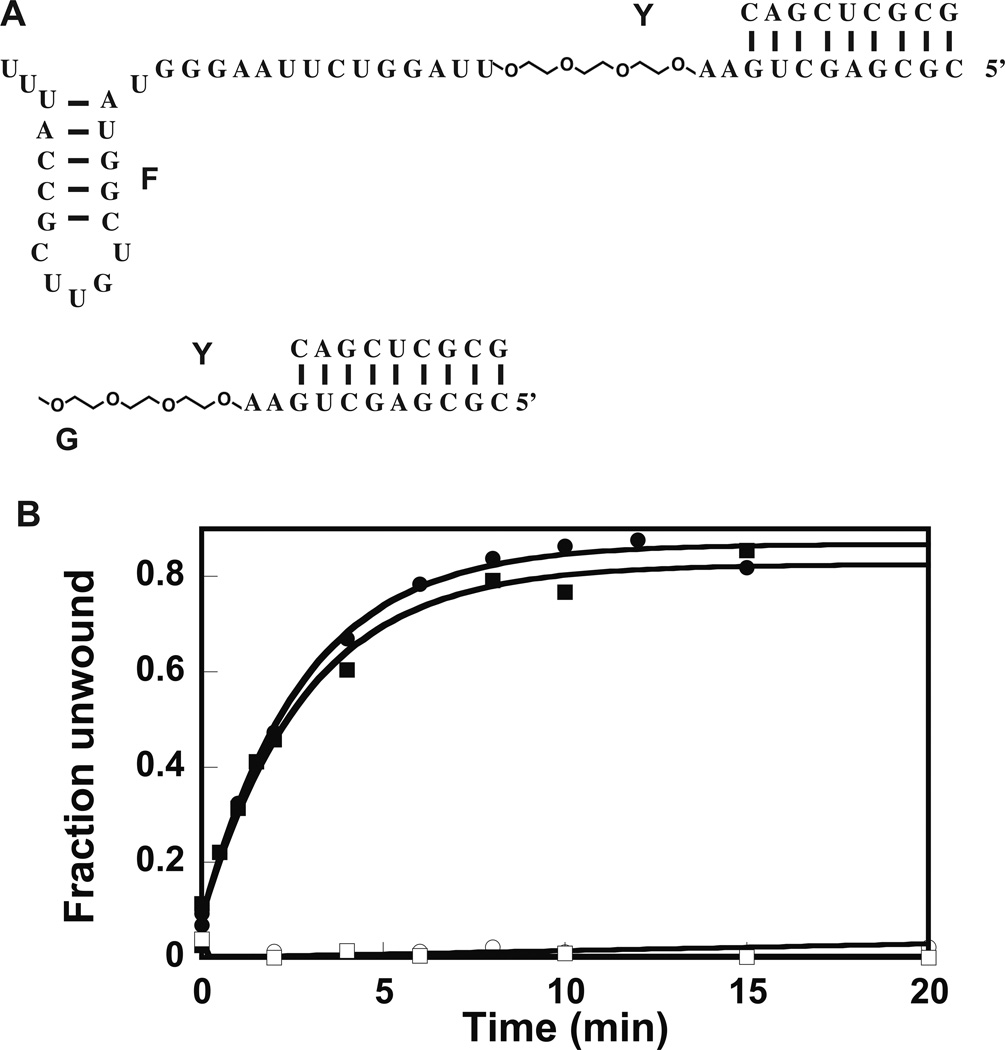
Figure 3.
Effect of PEG linker on the functional properties of DbpA. (A) The RNA–PEG chimera constructs used for these experiments. The 9-atom ethylene glycol linker spans 3 RNA bases. (B) Helicase activity of 23 amino acid extended and wild-type DbpA in presence of RNA–PEG chimeras. Legend: Filled square stimulation of the unwinding activity of wild-type DbpA by molecule F:Y; empty circle stimulation of helicase activity of wild-type DbpA by molecule G:Y; filled square stimulation of the unwinding activity of 23 amino acid extended DbpA by molecule F:Y; empty square stimulation of the unwinding activity of 23 amino acid extended DbpA by molecule G:Y. All the experiments shown here were performed at 600 nM protein. The helicase assay in presence of molecule G:Y was also performed at 2000 nM of protein (data not shown). Even at this much higher protein concentration, the construct G:Y could not support the helicase activity of wild-type or 23 amino acid extended DbpA. The data are representative of one experiment. The means of multiple experiments and the averages from the means are shown in Table 2.