Figure 1.
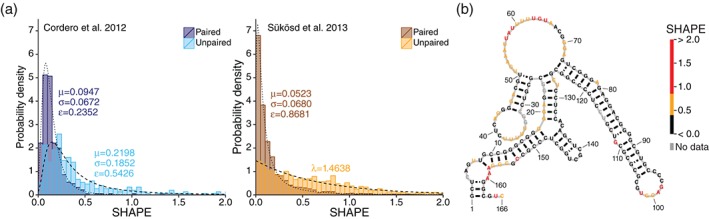
SHAPE reactivities distinguish between paired and unpaired nucleotides. (a) Distributions of SHAPE reactivities for paired and unpaired nucleotides recreated from Cordero et al.44 (left) and Sükösd et al.45 (right; data from Deigan et al.22). SHAPE reactivities for paired nucleotides follow a generalized extreme value distribution in both data sets. SHAPE reactivities for unpaired nucleotides follow a generalized extreme value distribution in Cordero et al. and an exponential distribution in Sükösd et al. In both data sets, the distributions for paired and unpaired nucleotides differ, signifying that SHAPE reactivities are drawn from multiple probability distributions. (b) Example of RNA structure (the RB1 5′ UTR from Kutchko et al.18) overlaid with SHAPE data. Red: high SHAPE; orange: medium SHAPE; black: low SHAPE; gray: no data. Most paired positions have low SHAPE and many unpaired positions have high SHAPE, but SHAPE does not completely distinguish between paired and unpaired nucleotides, in part because this RNA forms multiple conformations.
