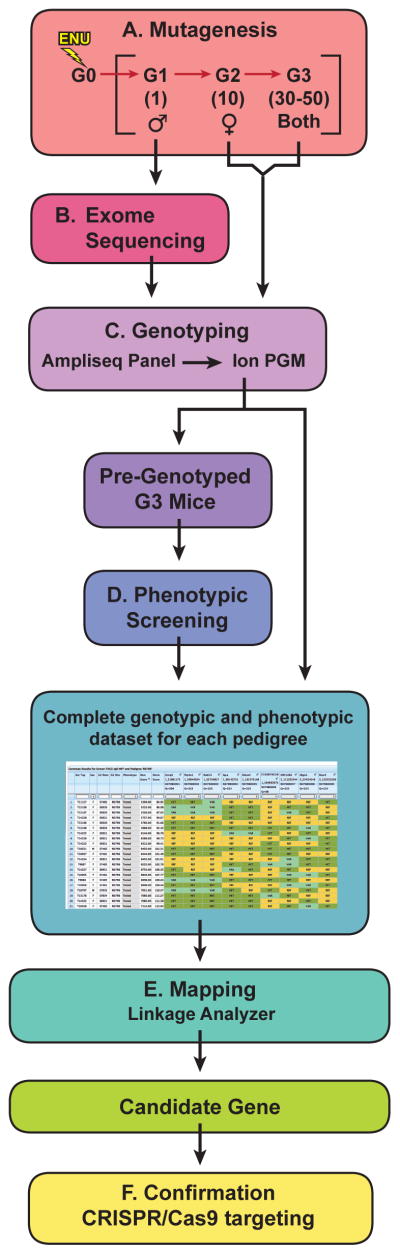
Figure 2.
Pipeline for mutagenesis, screening, and mapping to identify causative mutations in real time. (A) Male C57BL/6J mice mutagenized with ENU are bred to produce 30–50 third generation (G3) mice carrying mutations in homozygous and heterozygous state. (B) The G1 male founder of each pedigree is subjected to exome sequencing, and (C) data are used to generate Ampliseq panel primers for amplification of mutated loci from G2 and G3 mouse DNA, followed by Ion PGM 200-bp sequencing. Genotyping data are uploaded to the database prior to (D) phenotypic screening. Quantitative phenotype data are also entered into the database and used with genotype data for (E) mapping by Linkage Analyzer, a program developed in the lab. (F) Confirmation of candidate genes depends on duplication of the mutant phenotype by a second allele, which may be generated by CRISPR/Cas9 targeting.