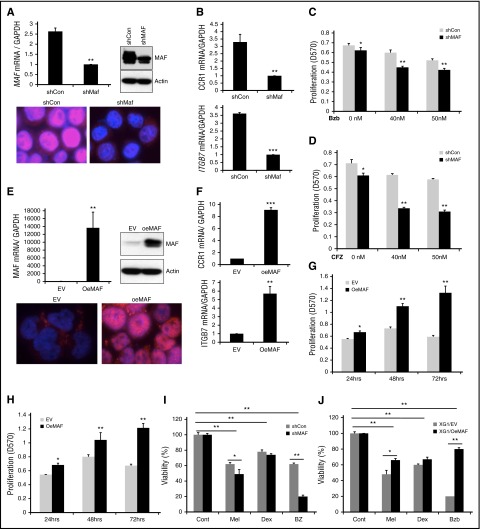
Figure 5.
Manipulation of MAF expression led to alteration in sensitivity of MM cells to PIs. 8226 cells were infected with lentiviral containing shRNA specific to MAF gene (shMAF) or shRNA containing scramble sequences (shCon) for 48 hours. The level of MAF mRNA in shMAF and shCon cells was determined by RT-qPCR. Results are presented as mean ± SE (n = 3). **P < .01 for shMAF vs shCon. The level of MAF protein in whole-cell lysates was determined by immunoblotting analysis. MAF protein expression in both cytoplasm and nuclei of shMAF and shCon cells was analyzed by immunofluorescent staining (A). The mRNA levels of CCR1 and ITGB7 in shMAF and shCon cells were measured by RT-qPCR analysis (B). Results are presented as mean ± SE (n = 3). **P < .01 or ***P < .001 for shMaf vs shCon. shMAF and shCon cells were treated with the indicated concentrations of Bzb (C) or CFZ (D) for 48 hours, and proliferation measured by MTT assay. Results are presented as mean ± SE (n = 4). Data are representative of 3 separate experiments. *P < .01 vs control. ***P < .001 vs control. Total RNA was obtained from XG1EV and XG1OeMAF and the levels of MAF mRNA (E), ITGB7 and CCR1 (F) was determined by RT-qPCR analysis. Results are presented as mean ± SE (n=3). **P < 0 .01 or ***P < 0 .001 for XG1EV vs XG1OeMAF. The levels of MAF protein in whole-cell lysate or in the cytoplasm and nucleus were determined (E). XG1OeMAF and XG1EV cells were treated with 20 nM Bzb (G) or CFZ (H) for indicated time points, and proliferation measured by MTT assay. Results are presented as mean ± SE (n = 4). Data are representative of 3 separate experiments. *P < .01 vs control. **P < .001 vs control. 8226/shMAF and 8226/shCon (I) or XG1OeMAF and XG1EV (J) cells were treated with or without Mel (50 μM), Bzb (40 nM), or Dex (250 μM) for 24 hours. Cell viability was measured by MTT assay. Results are presented as the percentage of alive cells relative to untreated cells. Each experiment was repeated n = 6 and reported as the mean value ± standard deviation (SD). *P < 0 .01 shMAF vs shCon; **P < 0 .001 shMAF vs shCon; **P < 0 .001 untreated vs treated shCon or untreated vs treated shMAF (I) and *P < 0 .01 XG1OeMAF vs XG1EV; **P < 0 .001 XG1OeMAF vs XG1EV; **P < 0 .001 untreated vs treated XG1EV or untreated vs treated XG1OeMAF (J).