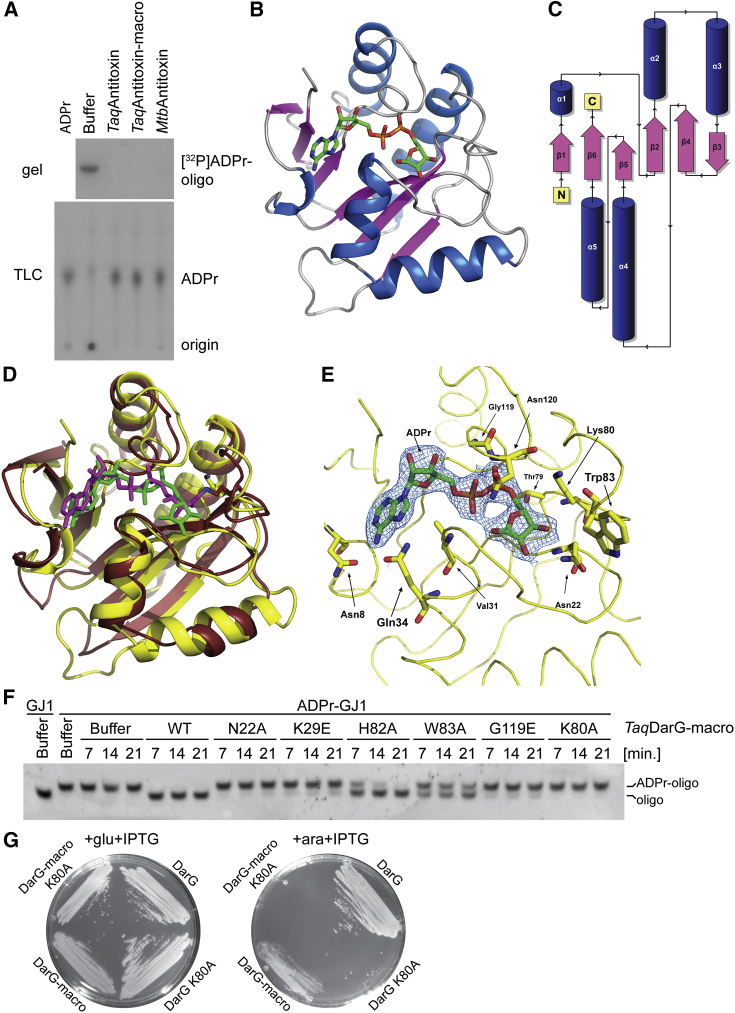
Figure 3.
Antitoxin Macrodomain De-ADP-Ribosylates DarT-ADP-Ribosylated Oligonucleotides
(A) Autoradiographs of denaturing polyacrylamide gel (top) or TLC plates (bottom) separating antitoxin reactions with 32P-NAD+ ADP-ribosylated oligonucleotide as substrate. Macro, macrodomain construct. ADPr standard reaction corresponds to poly-ADPr glycohydrolase-treated PARP1 reaction.
(B) Orthogonal view of TaqDarG-macro (cartoon) bound to ADPr (sticks).
(C) Topological diagram of the DarG macrodomain structures.
(D) Structural comparisons between TaqDarG-macro (yellow cartoon) bound to ADPr (green sticks) showing Lys80 (yellow sticks) and TARG1 (maroon cartoon; PDB: 4J5S) showing a covalent lysyl-ADPr adduct (magenta and maroon sticks).
(E) Close up of the active site of TaqDarG-macro showing the residues involved in ADPr binding. The ADPr ligand is shown with its 2Fo-Fc electron density contoured at 1σ.
(F) UV detection of ethidium bromide-stained denaturing polyacrylamide gel separating de-ADP-ribosylation reactions of TaqDarT ADP-ribosylated oligonucleotide by different TaqDarG-macro mutants. Reaction time in minutes is indicated at the top. Unmodified and ADP-ribosylated oligonucleotides were used as markers of migration.
(G) Images of bacterial growth at room temperature of BL21(DE3) with pBAD TaqDarT and pET vector encoding TaqDarG, DarG K80A, DarG-macro, or DarG-macro K80A. Plates were supplemented with glucose and IPTG for induction of expression from pET vector, or arabinose and IPTG for expression from both pET and pBAD vectors.