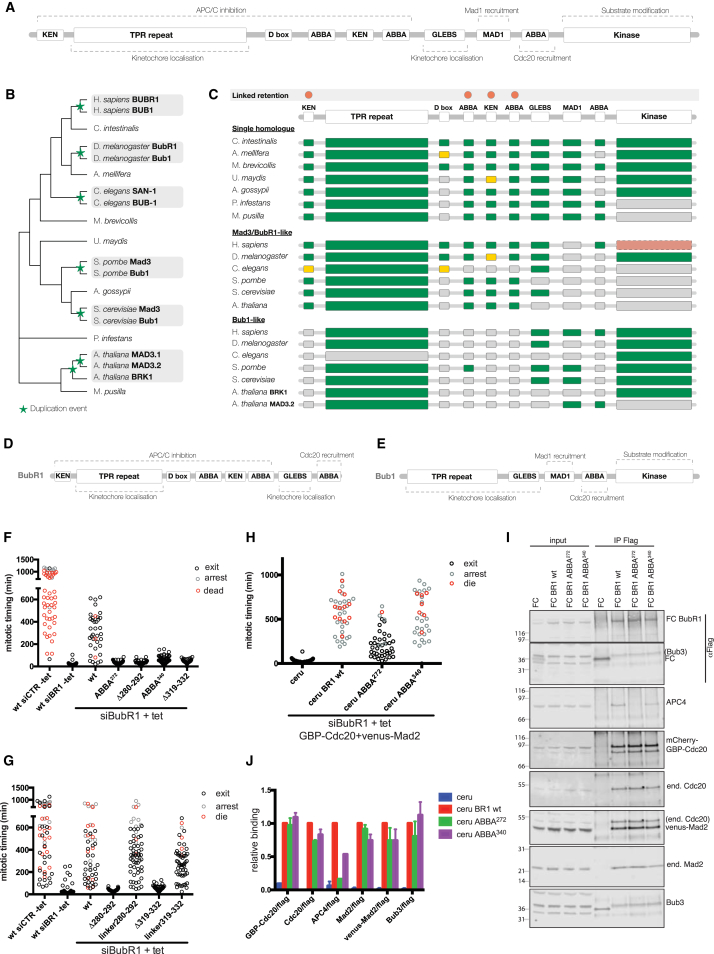
Figure 4.
The ABBA272-KEN2-ABBA340 Module Is Conserved through Evolution
(A) Predicted pre-duplication architecture of the ancestral protein to the human Bub1-like proteins—BubR1 and Bub1—based on Ciona intestinalis that has a single Bub1-like protein. The ancestral Bub1-like protein likely consisted of: an N-terminal KEN box pseudo-substrate domain; a TPR domain that stabilized the interaction between the N-terminal KEN and Cdc20 in the MCC (Chao et al., 2012) and bound to KNL1 to promote kinetochore recruitment (Krenn et al., 2012); a Bub3-binding motif (Wang et al., 2001); a MAD1 recruitment module (Klebig et al., 2009); an ABBA motif that promoted Cdc20 kinetochore recruitment (Di Fiore et al., 2015); and a C-terminal kinase domain (Bolanos-Garcia and Blundell, 2011, Suijkerbuijk et al., 2012). The region between the TPR domain and GLEBS motif also contained the putative ABBA motifs, a KEN box, and a D-box.
(B) An evolutionary tree of the Bub1-like proteins in selected eukaryotic species. The tree contains two distinct classes of species: (1) those where a duplication event has resulted in two or more Bub1-like proteins (light gray boxes) and (2) those where a single Bub1-like protein is present. The position of these classes in the tree was chosen to define points of Bub1-like protein duplication (green star).
(C) The modular architecture of the species in (B) grouped by (1) Bub1-like proteins from single homolog species, (2) BubR1-like proteins from multiple homolog species, and (3) Bub1-like proteins from multiple homolog species. Architecture shows the retained modules (green), inconclusively retained modules (yellow), retained modules characterized as non-functional (red), and potentially absent modules (gray). The modules that, post-duplication, have been simultaneously retained in BubR1 and lost in Bub1 independently on multiple occasions are marked above the architecture by red circles. Search details are shown in Table S1.
(D and E) Architecture of the functional modules in the human BubR1 (D) and human Bub1 (E) showing the role of the retained modules post-subfunctionalization. See Figure S3 and http://slim.ucd.ie/abbakenabba/ for alignments.
(F) HeLa FRT/TO cell lines stably expressing siRNA-resistant, FLAG-mRuby-tagged, WT or mutant BubR1 from a tetracycline (tet)-inducible promoter were transfected with control siRNA or siRNA against BubR1 (siBubR1), and nocodazole was added at the beginning of filming. Mitotic timing was measured with or without addition of tetracycline. The mitotic timing of WT BubR1 and ABBA272 or ABBA340 mutants was compared to deletion mutants in the inter-motif region. At least 50 cells per condition were analyzed. Results representative of three independent experiments.
(G) HeLa FRT/TO cell lines expressing the indicated BubR1 proteins were treated as in (F) and the mitotic timings compared. At least 50 cells per condition were analyzed. Results representative of three independent experiments.
(H) HeLa FRT/TO cell lines stably expressing siRNA-resistant, FLAG-Cerulean (FC) BubR1 WT or mutant from a tetracycline-inducible promoter were transfected with siRNA against BubR1 together with plasmids expressing mCherry-GBP-Cdc20 and Venus-Mad2. Mitotic timing was measured after addition of tetracycline. At least 35 cells per condition were analyzed. Results are representative of three independent experiments.
(I) HeLa FRT/TO cell lines stably expressing inducible siRNA-resistant, FLAG-Cerulean-BubR1 WT or mutants were transfected as for (H), and anti-FLAG immunoprecipitations from nocodazole-arrested cells were analyzed by immunoblotting and visualized on a LI-COR Odyssey scanner.
(J) The mean and SEM of protein levels from two independent experiments in (I). A third experiment was consistent with these results, but the siRNA did not deplete endogenous BubR1 to the same extent so these results were not included in the calculations.