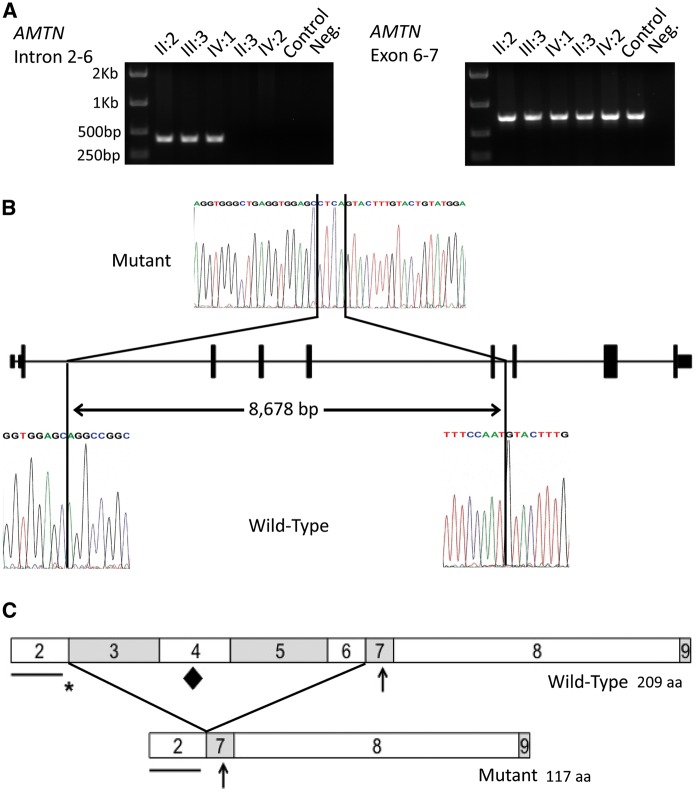
Figure 2.
Genotyping of the mutation, identification of the deletion breakpoints and a schematic diagram of the predicted effect of the deletion on the AMTN protein. (A) PCR analysis of the heterozygous deletion of exons 3 to 6 of AMTN in the members of the family investigated. Amplification using primers spanning introns 3 to 6, designed to produce a product of 9,073 bp for the wild-type allele, produced a product of 399 bp in affected individuals confirming a deletion encompassing exons 3 to 6. Amplification of exons 6 to 7 was observed in all individuals. (B) Sanger sequencing electropherograms of mutant and wild-type alleles to identify the deletion breakpoints. Comparison of the mutant sequence with a control sequence revealed a deletion of 8,678 bp spanning exons 3 to 6 of AMTN and an insertion of 4 bp. (C) Schematic diagrams of the wild-type and the predicted mutant AMTN protein structures. The predicted mutant AMTN protein lacks the amino acid sequence encoded by exons 3 to 6, resulting in the loss of 92 amino acids. The contribution of each exon to the protein is signified by the boxes labelled with the number corresponding to the exon number. Important motifs are labelled with symbols as follows: the line shows the position of the putative signal peptide, encoded by exon 2, the asterisk shows the position of the LPQ motif, encoded by exons 2 and 3, the diamond shows the position of the IPLT motif, encoded by exon 4, predicted to be an O-glycosylation site. The arrow shows the position of the SXE phosphorylation motif, encoded by exon 7.