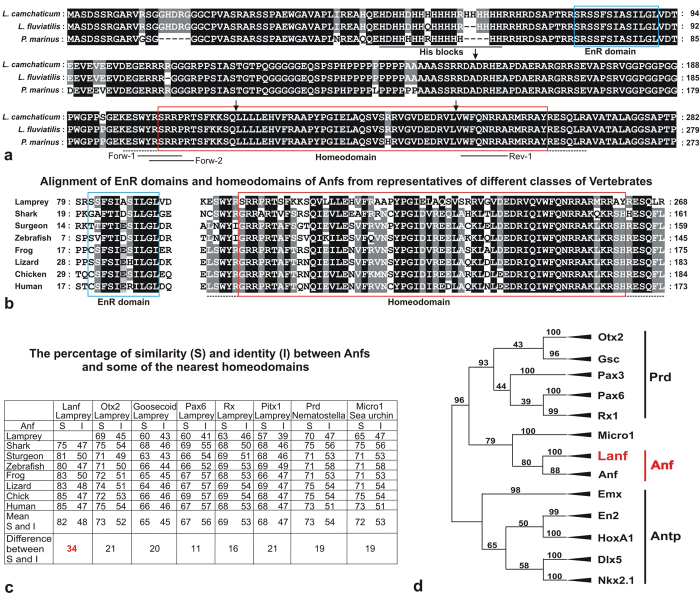
Figure 1. Sequences of lamprey Anfs.
(a) Alignment of Lanfs from L. camtschaticum, L. fluviatilis and P. marinus (GenBank: KX245018, KX245019, KX245020). Homeodomains and the engrailed repressor domain (EnR) are indicated in red and blue frames, respectively; histidine-rich blocks are underlined with a solid black line; conservative motifs flanking the homeodomain are underlined with a dotted line; arrows indicate the positions of introns; the positions of the degenerate primers used for RT-PCR during Lanf cloning are indicated with solid lines. (b) Alignment of the EnR domains and homeodomains of L. camtschaticum Lanf with those of Anfs from different classes of vertebrates. (c) The percentages of similarity (S) and identity (I) between Anfs and some of the homeodomains of other types. The mean difference between the similarity (S) and identity (I) of the Lanf homeodomain with homeodomains of other Anfs (bottom line, highlighted by red) is much greater than the analogous values calculated for Lanf and other types of homeodomains (bottom line, in black). This fact indicates conservation of Lanf function during evolution. (d) Scheme of the neighbour-joining phylogenetic tree of Anf homeodomains and homeodomains of Antp- and Prd-class proteins (full trees constructed via the neighbour-joining and maximum likelihood methods are shown in Fig. S2, Fig. S3 and Fig. S4).