Abstract
Transition metals are essential constituents of all living organisms, playing crucial structural and catalytic parts in many enzymes and transcription factors. However, transition metals can also be toxic when present in excess. Their uptake and efflux rates must therefore be carefully controlled by biological systems. In this chapter, we summarize the current knowledge about uptake and efflux systems in Mycobacterium tuberculosis for mainly three of these metals, namely iron, zinc and copper. We also propose questions fur future research in the field of metallobiology of host-pathogen interactions in tuberculosis.
The battle for iron
Iron is absolutely required for the life of most organisms, including mycobacteria. Iron is incorporated into proteins, either as a mono or binuclear species, or as part of heme groups or iron-sulfur clusters. Iron undergoes reversible changes in its oxidation state oscillating between the oxidized ferric (Fe+3) and the reduced ferrous (Fe+2) forms. In addition, depending on the local ligand environment, iron-containing compounds exhibit a wide range of oxidation-reduction potentials. These unique properties make this metal a very versatile prosthetic component as biocatalyst and electro-carrier in essential cellular pathways including respiration, the trichloroacetic acid (TCA) cycle, oxygen transport, gene regulation, defense against oxidative stress and DNA biosynthesis (1).
Iron is the fourth most plentiful element in the earth crust. Before oxygenic photosynthesis it was found in its soluble ferrous form (solubility 0.1 M at pH 7.0); however, introduction of oxygen into the atmosphere caused a switch to the ferric form, which is insoluble as ferric hydroxide. In consequence, free iron became extremely scarce (solubility 10−18 M at pH 7.0). In host tissues, the concentration of this metal is lowered even further as Fe(III) is sequestered by iron-binding proteins such as transferrin, lactoferritin and ferritin (2, 3). In addition, the host produces proteins that either efflux iron from intracellular microbial compartments (NRAMP1), or bind heme and haemoglobin (e.g. hemopexin and haptoglobin) and reduce the availability of heme as iron source (2, 4, 5). Thus, iron starvation in the host is a serious threat for infecting bacteria, and has been recognized as such since decades ago (3). Pathogens are able to survive and multiply in the host in part because they have evolved numerous and often redundant high-affinity iron acquisition mechanisms, including: a) acquisition of iron directly from host iron-binding proteins (e.g. transferrin and lactoferrin) by using receptor-mediated transport systems; b) uptake and utilization of heme; c) solubilization of ferric oxides by reduction of ferric iron and transport of soluble ferrous iron, and d) production of ferric iron chelators (siderophores) in conjunction with siderophore-based transport systems. M. tuberculosis obtains iron by producing siderophores and it also has the capacity to utilize heme as iron source, in a siderophore-independent manner.
Siderophore-based iron acquisition in M. tuberculosis
M. tuberculosis produces mycobactin, a cell-associated, lipophilic siderophore and a soluble amphiphilic variant of it named carboxymycobactin (6). M. tuberculosis does not produce or utilize exochelin, the peptidic siderophore synthesized by non-pathogenic mycobacteria such as Mycobacterium smegmatis. Mycobactin and carboxymycobactin are composed of a hydroxyaromatic acid, an oxazoline moiety, a β-hydroxy acid and two ε-N-hydroxylysines (Figure 1). Genomic and biochemical analysis indicate the mbt-1 gene cluster (mbt-IABCDEFG) which encodes for salicylate synthesis (MbtI), the L-Lys hydroxylase (MbtG) and the hybrid non-ribosomal peptide synthase/polyketide synthase (MbtA-F) is involved in the assembly of the siderophore core (7–9). Mycobactin and carboxymycobactin differ mainly in an acyl group attached to the central L-Lys residue. Mycobactin has a long fatty acyl chain (10 to 21 carbons) that is capped by a methyl group, whereas carboxymycobactin has a shorter acyl chain (2 to 9 carbons) that is capped with either a carboxylate or methyl ester (10, 11). The presence of the long acyl chain makes mycobactin very hydrophobic and ensures retention within or in close proximity to the cytoplasmic membrane. Mycobactin may mediate uptake of iron donated by amphiphilic molecules that can penetrate the cell wall, for instance carboxymycobactin and acinetoferrin (12, 13). Carboxymycobactin being more polar than mycobactin, it is water soluble and exported to the extracellular medium. The process of carboxymycobactin export seems to be coupled -in an unknown way- to its synthesis and depends on two redundant systems composed of the MmpL4 and MmpL5 transporters and their associated proteins MmpS4 and MmpS5 (14). These transport systems are postulated to mediate export of carboxymycobactin into the periplasm. However, the hypothethical outer membrane protein that mediates release of carboxymycobactin into the extracellular medium is unknown (Figure 2). M. tuberculosis mutants unable to synthesize or export siderophores are drastically attenuated in a mouse model of tuberculosis infection, underscoring the importance of efficient iron acquisition for propagation of M. tuberculosis (14).
Figure 1.
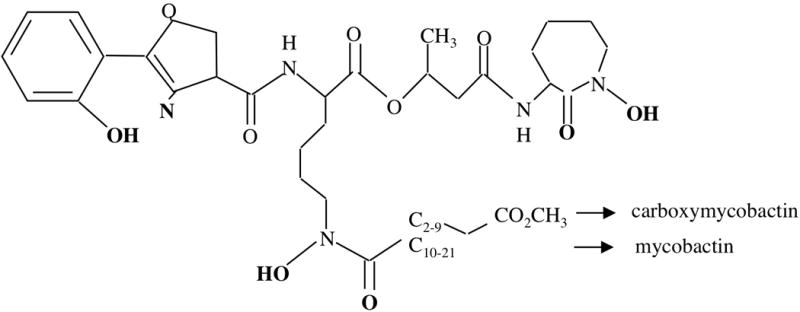
Carboxymycobactin and mycobactin share a common core structure but differ in the length of the alkyl substitution which determines their polarity and hence solubility. The groups involved in binding of Fe(III) are indicated in bold.
Figure 2.
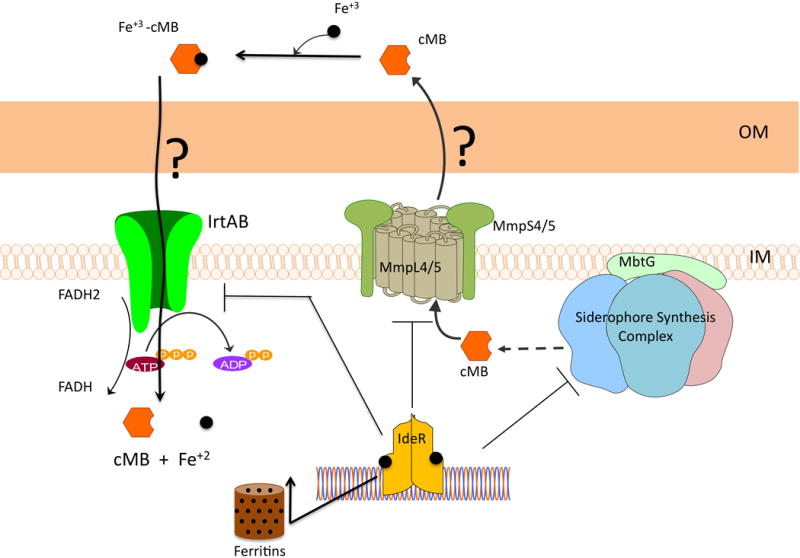
When experiencing iron limitation, M. tuberculosis produces carboxymycobactin (cMB) and mycobactin (MB). Mb remains cell associated although, the precise location is not clear. cMB is secreted by a process dependent on the membrane proteins MmpL4 and MmpL5 and requiring the MmpS4 and MmpS5 membrane-associated proteins which function together with their cognate MmpL proteins. Protein(s) that mediate export of cMB across the outer membrane remain to be discovered. Once secreted, cMB chelates Fe+3 and possibly requires an outer membrane and periplasmic protein to reach the IrtAB importer in the inner membrane. In the cytosol the FAD binding domain of IrtA may reduce ferric iron to ferrous iron and dissociate the iron-siderophore complex. Released ferrous iron can be utilized and stored in ferritins. Excess iron binds to the regulator IdeR and activates its DNA binding activity. Binding of IdeR to the promoters of siderophore synthesis, secretion and transport represses the expression of those genes turning off iron uptake. Meanwhile IdeR-Fe+2 binding to the promoters of ferritins (ferritin and bacterioferritin) turns on iron storage thereby, preventing iron mediated toxicity and maintaining iron homeostasis.
In the extracellular environment carboxymycobactin avidly captures ferric iron. Ferric-carboxymycobactin can slowly transfer iron to mycobactin (12), or deliver this metal via the iron-regulated transporter IrtAB. IrtAB is an ABC-type transporter synthesized in cells experiencing iron limitation, necessary for Fe+3-carboxymycobactin uptake. IrtAB mutants are iron deficient and fail to replicate normally in macrophages and in mice (15). Interestingly, the amino terminal domain of the IrtA protein is located in the cytoplasm and has a functional FAD (flavin adenine dinucleotide) binding motif. Mutations that prevent FAD binding affect assimilation of iron imported by IrtAB. Since a common mechanism to dissociate iron-siderophore complexes is reduction of ferric iron to ferrous iron by cytoplasmic flavin reductases, it is possible that the amino terminal domain of IrtA functions as a FAD dependent ferric-reductase that mediates the release of ferrous iron from imported Fe+3-carboxymycobactin (16). Thus, IrtAB might couple iron transport and assimilation in M. tuberculosis (Figure 2).
Considering the double membrane structure of the mycobacterial cell envelope, it is likely that Fe+3-carboxymycobactin uptake is facilitated by siderophore binding proteins in the outer membrane and the periplasm. These proteins, however, are yet to be identified. The mycobacterial type VII secretion system, Esx-3, is induced under low iron conditions and has been shown to be necessary for growth of M. tuberculosis and M. bovis BCG in iron-deficient medium and for utilization of exogenously added Fe+3-carboxymycobactin (17, 18). These findings suggest that directly or indirectly, components of the Esx-3 system may contribute to Fe+3-carboxymycobactin uptake.
Heme utilization
Many pathogens have evolved strategies to obtain iron from heme, which is the most abundant source of iron in mammals. Bacteria can obtain heme using outer membrane receptors and periplasmic binding protein-dependent ABC transporters specific for heme, or they synthesize and secrete specialized proteins (hemophores) able to sequester heme and deliver it to a specific outer membrane receptor. M. tuberculosis is able to obtain iron from heme in the absence of siderophores (19). A genetic region encoding a secreted heme-binding protein (hemophore) and two membrane transporters is necessary for normal iron acquisition from heme and hemoglobin (20). Once internalized, heme has to be degraded to release iron. This function is usually performed by heme oxygenases that degrade heme into iron, a tetrapyrrole product and carbon monoxide (CO). In M. tuberculosis this role might be performed by the enzyme MhuD, a homolog of the Staphylococcus aureus heme oxygenases IsdG and IsdI. MhuD degrades heme in a unique way: it releases iron and a tetrapyrrole product named mycobilin but without CO generation (21). Presently, the role of heme uptake in the pathogenesis of M. tuberculosis is unknown. Studies in animal models will determine the relevance of heme utilization in M. tuberculosis virulence.
Iron storage
The synthesis of iron storage proteins (ferritins) is central to iron homeostasis in most aerobic organisms and necessary for virulence of many pathogens. Ferritin subunits form a hollow sphere where up to 4,500 atoms of iron can be sequestered as mineral, after being oxidized to Fe+3 at a ferroxidase center (22). Some bacteria and fungi synthesize ferritin-like proteins containing haem b known as bacterioferritins. M. tuberculosis has a bacterioferritin (BfrA) and a ferritin (BfrB). The crystal structure of these proteins show the typical architecture of the ferritin superfamily of a cage-like hollow shell formed by 24 monomers with the characteristic fold of a four-helical bundle containing the ferroxidase catalytic center, and in bacterioferritin a heme group in each subunit-pair interface (23, 24). Analyses of single deletion mutants of M. tuberculosis, showed that BfrA and BfrB are not functionally redundant. Deletion of bfrB drastically altered iron homeostasis whereas no obvious defects were detected in a bfrA deleted mutant (25). Iron stored by BfrB seems to be M. tuberculosis’ preferred reserve to overcome iron deficiency. In addition, M. tuberculosis lacking BfrB is highly sensitive to peroxide and antibiotic generated oxidative stress when cultured in iron rich media. This indicates that BfrB is required to prevent excess free iron from catalyzing the generation of toxic reactive oxygen species (25). The significance of proper iron storage in pathogenesis of M. tuberculosis has been demonstrated in animal models of infection. A mutant lacking bfrB is unable to persist in the lungs of mice and establish infection in the liver (25). Furthermore, a double bfrA/bfrB mutant is strongly attenuated in a guinea pig model of tuberculosis infection (26).
In addition to BfrA and BfrB, M. tuberculosis possesses a histone-like DNA binding protein (MDP1) that captures iron and also has ferroxidase activity. MDP1 may protect DNA by preventing the local generation of reactive oxygen radicals (27).
Regulation of iron metabolism
Iron can be very toxic because it catalyzes the generation of reactive oxygen species from normal products of aerobic respiration via the Harber-Weiss and Fenton reactions. Reactive oxygen species can damage most cellular components including DNA, lipids and proteins. For this reason, aerobic organisms must tightly control intracellular iron levels. In bacteria, this control is generally achieved by regulating the uptake, utilization and storage of this metal. As other prokaryotes, M. tuberculosis regulates iron metabolism at the level of gene transcription. It induces the expression of iron uptake genes under iron deficiency and up-regulates iron storage and oxidative stress defense genes when iron is readily available (28). M. tuberculosis achieves the delicate balance between the requirement for iron and its toxicity through the function of the iron-dependent regulator IdeR. IdeR is a metal and DNA binding protein, closely related to the Corynebacterium diphtheriae regulator of iron metabolism and toxin production DtxR (29). The structure of IdeR revealed two metal binding sites and three distinct functional domains. The amino-terminal containing a helix-turn-helix DNA binding motif, a dimerization domain that also bears most of the metal binding residues, and the carboxy-terminal domain characterized by adopting an SH3 (Src homology domain 3)-like folding, suggesting possible interactions with other proteins (30). Metal binding stabilizes dimer formation (31) and activates DNA binding. As two dimers, IdeR binds to both faces of the DNA at a unique 19-bp inverted repeat sequence, the “iron box” (TTAGGTTAGGCTAACCTAA), present in the promoter of iron-regulated genes, thereby modulating their transcription (32). Disruption of the ideR gene in M. tuberculosis is only possible in the presence of a second copy of the gene or when suppressor mutations arise. This indicates that IdeR is essential in M. tuberculosis (28). Approximately 150 genes respond to changes in iron availability in M. tuberculosis. IdeR controls the expression of about one third of those genes including: the siderophore synthesis and export genes, the siderophore transporter encoding genes irtA and irtB, genes in the esx-3 cluster and the iron storage genes bfrA and bfrB (28, 32). IdeR and Fe+2 turn off iron acquisition and turn-on iron storage (Figure 2). These opposite effects of IdeR as repressor of iron uptake and activator of iron storage can be understood by considering the position of the iron box in repressed and activated promoters. Iron boxes on IdeR-repressed genes overlap the -10 region or the transcriptional start site; consequently, binding of IdeR to the iron box blocks access of the RNA polymerase and inhibits transcription of those genes. In the promoters of bfrA and bfrB, tandem iron boxes are located farther upstream (100–106 bp) from the transcriptional start site, suggesting a mechanism of activation by which IdeR-Fe+2 bound to these sites enhances access of the RNA polymerase to the promoter and initiation of transcription. In view of the strong attenuation of iron storage mutants in vivo (25), it is likely that IdeR-mediated activation of iron storage is essential for growth of M. tuberculosis during infection.
Zinc and copper: never too little or too much
Zinc and copper play vital functions in biological systems. The chemical properties of zinc, e.g. its Lewis acidity, coordination geometry and rapid ligand exchange, allow it to form stable complexes with enzymes and proteins where it functions in catalysis or as a structural factor. The majority of zinc containing enzymes catalyze hydrolysis or related transfer reactions, some of which being essential for cell viability. The number of Zn containing proteins identified in mycobacteria has increased significantly as more protein structures are resolved. Zn is part of M. tuberculosis zinc-metallopeptidases (33, 34), carbonic anhydrase (35), fructose biphosphate aldolase Fba (36), the helicase RqlH (37), the cytidine deaminase Cda (38), the MshC ligase involved in mycothiol biosynthesis (39), the 2C-methyl-D-erythritol-2,4-cyclodiphosphate synthase IspF (40), the 2-isopropylmalate synthase LeuA involved in leucine biosynthesis (41), the superoxide dismutase (SOD) SodC (42, 43), the Esx-3 substrate EsxG-EsxH complex (44), the inositol 1-phosphate synthase (45), the RecA intein (46) and several more.
Because of its fast interconversion of Cu+ and Cu2+, copper is involved in several essential biochemical processes, such as oxygen-dependent electron transport reactions. In M. tuberculosis, at least two enzymes require copper as co-factor, namely the SOD SodC (42, 43) and the cytochrome c oxidase subunits CtaC and CtaD.
Bioavailable levels of zinc are sufficiently low that most microbes have evolved high affinity transport systems to capture this metal. Bacterial zinc transporters are generally ABC transporters consisting of a periplasmic binding protein, a membrane permease and an ATPase. The periplasmic binding protein, which usually have a central His-, Asp-, and Glu-rich region, seems to allow specificity for zinc over manganese and other cations (47). Proteins involved in zinc import in mycobacteria have yet to be discovered. Regarding copper, as in most bacterial species, uptake systems for this metal have not been identified in M. tuberculosis.
Metallobiology of zinc and copper in M. tuberculosis recently provided insights into novel host defense mechanisms against bacterial infection involving intoxication by metal ions. To resist potential intoxication by metal ions, microbes express a range of metal efflux pumps and transporters belonging to three main families: heavy metal efflux members of the resistance–nodulation–cell division superfamily (HME-RND), the cation diffusion facilitators (CDF) family, and the P-type ATPase family (48). A set of recent studies strikingly reported that some of these efflux systems are required for microbial virulence in various bacterial species, including M. tuberculosis, in order to resist newly described immune mechanisms relying on metal poisoning of microbes inside host cells.
In the M. tuberculosis genome, no putative heavy metal efflux system of the HME-RND family is detected, while one putative CDF transporter (Rv2025c) and 12 putative P-type ATPase members (CtpA-J, CtpV and KdpB) are present (48, 49). The exact substrate specificity of these transporters is not known, and is mostly inferred from indirect evidence such as similarity to known transporters and presence of conserved metal-binding motifs. For instance, the transcriptional regulator CmtR/Rv1994c, present in operon with the P-ATPase CtpG, responds to cadmium and lead (50) to alleviate ctpG transcriptional repression, suggesting that CtpG can efflux these two heavy metal cations; similarly, the ability of NmtR/Rv3744 to respond to nickel and cobalt, and to bind the promoter region of the neighbor gene ctpJ/nmtA/Rv3743c to repress its expression in metal-free conditions (51) again suggests CtpJ may efflux nickel and cobalt. Finally, the recent findings that M. tuberculosis mutants inactivated in ctpV and ctpC, are highly sensitive to copper and zinc, respectively (52, 53), strongly suggest these two P-ATPases may transport these metal ions, but again is not a proof of their metal selectivity. Biochemical characterization of these transporters in recombinant biological systems and in reconstituted liposomal fractions will be required in order to understand their function. In this context, a striking feature of three P-ATPase members in M. tuberculosis, namely CtpC, CtpG and CtpV, is the presence of a putative metallochaperone-encoding gene, namely and respectively Rv3269, Rv1993c, and Rv0968, upstream of the P-ATPase-encoding genes. The P-ATPase- and the metallochaperone-encoding genes seem to be expressed in operon. The function of these small proteins, predicted to be membrane bound, and exposing putative metal binding motifs (e.g. DDGHDH in Rv0968) in their C-terminal cytoplasmic part, is not known ; however it is most tempting to speculate that they may play a key part in metal selectivity and transport mechanism of their cognate P-ATPase, as recently demonstrated for a similar transport system in Streptococcus pneumoniae (54).
A role for P-ATPase-mediated metal detoxification in M. tuberculosis has been recently suggested by several independent reports. In particular M. tuberculosis mutants inactivated in the P-ATPase-encoding genes ctpV and ctpC were shown to be impaired in their ability to proliferate in model animals and/or host macrophages (52, 53). In a guinea pig model, Ward et al. reported that lung colonization by M. tuberculosis ΔctpV was reduced by ≈1-log10 three weeks after inoculation, as compared to the wild-type strain, and full virulence of the mutant was restored upon genetic complementation (53). The same authors reported a similar observation in mice, where the survival rate of animals infected with the mutant strain was increased by 16 weeks, as compared to those infected with the wild-type strain, although unlike in guinea pigs, no CFU difference were noticed in the mouse lungs. In both animal models, lung granulomatous inflammation was severely reduced in animals infected with the ΔctpV mutant, as compared to the wild-type strain. Although no direct demonstration has been provided yet regarding the metal selectivity of CtpV, it is most likely that this P-ATPase effluxes copper because i/ the ctpV gene is induced by copper (55, 56), ii/ the CtpV protein contains typical motifs of the P1B1 family of copper-transporting P-ATPases (Table 1), iii/ the ctpV gene is encoded in operon with the copper-responsive transcriptional repressor CsoR (57), and most importantly, iv/ the ΔctpV mutant is highly sensitive to copper in vitro (53). These results suggesting that M. tuberculosis is facing copper intoxication in vivo during infection were further strengthened by another report where it was shown that the outer membrane channel protein Rv1698/MctB is also required for both copper detoxification in vitro and for full virulence in vivo in guinea pigs (58). It was thus proposed that copper accumulation inside the mycobacterial phagosome may account for the phenotype of the ΔctpV and ΔmctB mutants in vivo (59–61).
Table 1.
P-ATPases in M. tuberculosis
| Gene name | Group | Predicted substrate(s) | Comments | References |
|---|---|---|---|---|
| ctpA/Rv0092 | 1B1 | Cu+/Ag+ | N-term CxxC; C-term MxxSS | |
| ctpB/Rv0103c | 1B1 | Cu+/Ag+ | N-term CxxC; C-term MxxSS | |
| ctpC/Rv3270 | 1B? | Zn2+; possibly others | Putative metallochaperone Rv3269; M. tuberculosis mutant sensitive to zinc | (53) |
| ctpD/Rv1469 | 1B4 | Co2+ | C-term HEGxT; M. smegmatis homologue transports Co2+ | (84) |
| ctpE/ | 2A-like | Unknown | PEGL(P/V) motif found in calcium-transporting P-ATPases, such as SERCA | |
| ctpF/ | 2A | Ca2+ | PEGL(P/V) & Tm6-LWxNxxxd motifs found in calcium-transporting P-ATPases, such as SERCA | |
| ctpG/Rv1992c | 1B? | Possibly Cd2+/Pb2+ & others | Putative metallochaperone Rv1993c; repressed by Rv1994c/CmtR, unless Cd2+ or Pb2+ are present | (81) |
| ctpH/ | 2A-like | Unknown | Large N-term membrane spanning domain; Tm6-PEGl(P/V) motif found in calcium-transporting P-ATPases, such as SERCA | |
| ctpI/ | 2A-like | Unknown | Large N-term membrane spanning domain; Tm6-PEGL(P/V) motif found in calcium-transporting P-ATPases, such as SERCA | |
| ctpJ/Rv3743c | 1B4 | Co2+ | C-term HEGxT; repressed by Rv3744/NmtR, unless Ni2+ or Co2+ are present | (51) |
| ctpV/Rv0969 | 1B1 | Cu+ | C-term MxxSS; M. tuberculosis mutant sensitive to copper; putative metallochaperone Rv0968; in operon with copper-responsive regulator csoR/Rv0967 | (53, 56) |
| kdpB/Rv1030 | 1A | K+ | Homologous to many KdpB potassium transporters |
Phagosomal intoxication by copper has been suggested in other settings; in particular an elegant study conducted in Escherichia coli-infected macrophages, reported that copper enhances intracellular bacterial killing inside macrophages, and that an E. coli mutant inactivated in the copper efflux P-ATPase CopA is killed faster in macrophages than its wild-type counterpart, unless the eukaryotic copper transporter ATP7A, that traffics to phago-lysosomes, is silenced through iRNA (62). Although copper accumulation in the bacterial vacuole was not directly evidenced, this elegant study suggested for the first time that copper was an important mediator of microbial killing by immune cells and provided a mechanistic explanation for this phenomenon.
Regarding CtpC, we reported that genetic inactivation of this P-ATPase dramatically increases M. tuberculosis sensitivity to Zn2+, which strongly suggested CtpC might be involved in zinc efflux (52). However, a recent report suggested that CtpC may transport Mn2+over Zn2+, and that the hypersensitivity of the ctpC mutant to zinc may be due to an increased sensitivity to oxidative stress following impaired Mn2+ loading of the SOD SodA and possibly other detoxification systems (63). Inside macrophages, we showed that zinc accumulates within E. coli- or M. tuberculosis-containing phagosomes, and that bacterial strains impaired in resistance to zinc (a ΔzntA mutant in E. coli or a ΔctpC mutant in M. tuberculosis) are impaired in intracellular survival. In vivo attenuation of the M. tuberculosis ΔctpC mutant still has to be clearly established (52, 63).
The requirement of P-ATPase-mediated copper resistance systems in bacterial virulence has been documented in several bacterial species, including Listeria monocytogenes (64), Pseudomonas aeruginosa (65), S. pneumoniae (66), and Salmonella typhimurium (67). Several mechanisms have been proposed to explain copper ion toxicity. These mechanisms include Fenton chemistry and generation of hydroxyl radicals, although this was challenged by data showing that there is no accumulation of hydroxyl radicals in copper-exposed E. coli (68); degradation of iron-sulfur clusters in enzymes (69); and replacement of other metal ion cofactors, such as zinc ions, in proteins. The exact mechanism(s) of copper toxicity in M. tuberculosis remain to be identified.
The mechanism(s) of zinc ion toxicity may also include inactivation of iron-sulfur clusters (70), and inhibition of manganese uptake through transport competition in the bacterial perisplam (71). It was shown recently that P-ATPase-mediated copper export is required for copper supply to periplasmic Cu,Zn-SOD and resistance to oxidative stress in Salmonella enterica (72). Whether copper and zinc export through CtpV, CtpC and possibly other P-ATPase contributes to activation of the periplasmic Cu,Zn-SOD SodC, in M. tuberculosis remains to be evaluated.
In summary, it is clear that M. tuberculosis uses the P-ATPase CtpC and CtpV to thrive inside macrophages and resist poisoning by Zn2+ and Cu+. The function of the other M. tuberculosis P-ATPases, and their possible implication in mycobacterial virulence, remains to be understood. Equally important will be to understand the function of the putative metallochaperones associated to CtpC, CtpG and CtpV.
Regulation of metal uptake
As stated above, although necessary, zinc and other metal ions can also be toxic if present at too high concentration. For instance, zinc may interact with thiols or compete with other metals for protein binding, blocking essential reactions in the cells. Therefore, quantity of zinc inside the cells is carefully regulated, usually by calibrating uptake and export. The genome of M. tuberculosis contains two genes encoding transcriptional regulators of the Fur family, FurA and FurB. Structural and functional characterization of FurB revealed it to be a Zn+2-dependent repressor, hence, it has been renamed Zur (zinc uptake regulator) (73–75). Genes repressed by Zur-Zn+2 include the gene cluster encoding the Esx-3 secretion system, several ribosomal proteins, a protein similar to the Bacillus subtilis low-affinity zinc transporter YciC, and components of a putative ABC-type Zn+2/Mn+2 transport system (74). Disruption of the zur gene did not affect the ability of M. tuberculosis to replicate in mice suggesting that constitutive expression of Zur-regulated genes is not detrimental for M. tuberculosis in this model of infection (74).
The importance of sensing metal deficiency or excess is reflected in the multiple families of metalloregulatory proteins characterized in bacteria. These include: Fur, DtxR, MerR, SmtB/ArsR, CsoR, CopY and NikR. In general, these proteins are transcriptional regulators that sense specific metal ions via direct coordination. The DtxR, Fur and NikR family proteins primarily regulate genes required for metal uptake, whereas members of the other families regulate mainly metal efflux. Fur was first described as an iron-responsive repressor of iron transport in E. coli. Since then, numerous studies have revealed functional specialization within the Fur family and a great diversity in metal selectivity and biological function. The Fur family includes sensors of iron (Fur), zinc (Zur), manganese (Mur) and nickel (Nur). Some members of the family use metal catalyzed redox reactions to sense peroxide-mediated stress (Per) or heme (Irr).
M. tuberculosis has two Fur-like proteins, namely Zur (described before) and FurA. The FurA-encoding gene is located immediately upstream of katG, the gene encoding a catalase-peroxidase, a major virulence factor that it also activates the prodrug isoniazid. FurA and katG are co-transcribed from a common promoter upstream of furA. FurA auto-represses its expression and the expression of katG by binding to a unique sequence upstream of furA (76–78). FurA seems to have a very specialized biological role as no other genes regulated by FurA have been identified to date.
Two regulators of the DtxR family are present in M. tuberculosis: the iron-dependent regulator IdeR (described earlier) and SirR (for Staphyloccocal iron-regulated Repressor). In S. epidermidis SirR, in complex with Fe+2 or Mn+2, binds to a unique sequence in the promoter of an operon encoding for a putative iron transporter (79). The biological role of the SirR homologue in M. tuberculosis, however, has not been determined.
Other metalloregulators characterized in M. tuberculosis include: the Ni (II)/Co(II) specific repressors NmtR (51) and KmtR (80), the copper sensors CsoR (57) and RicR (55), the Cd(II)/Pb(II) sensor CmtR (81) and the Zn(II) responsive regulator encoded by the gene Rv2358 (82). In general they regulate the transcription of membrane transporters that mediate cytoplasmic efflux of potentially toxic metals, as mentioned above.
Future directions
Much remains to be understood regarding the mechanisms of transition metal uptake/efflux systems in M. tuberculosis, their regulation, and the biological impact of selective metal ion enrichment or depletion encountered in the mycobacterial phagosome inside host macrophages (59, 83). As mentioned above, the zinc and copper uptake systems still have to be identified in M. tuberculosis. Identification of the remaining components of the iron acquisition apparatus and a better understanding of the mechanisms that control iron sorting and assimilation in M. tuberculosis will reveal new possibilities of intervention. For instance, ways to starve M. tuberculosis for iron or alternatively, get it to intoxicate itself by corrupting its iron sensing mechanisms may help develop novel treatments. Future work should also aim at deciphering the exact metal specificity and biological function of the many M. tuberculosis P-ATPases, and the use innate immune cells make of metal ion withdrawal or intoxication to contain mycobacterial infection.
Acknowledgments
The authors received no specific funding for this work. The Neyrolles laboratory is supported by the Centre National de la Recherche Scientifique (CNRS), the European Union (7th Framework Pro-gramme), the Agence Nationale de la Recherche (ANR), the Fondation Mérieux and the Fondation pour la Recherche Médicale (FRM). Work from the Rodriguez’s laboratory discussed in this chapter was supported by NIH research grant AI44856 (GMR).
References
- 1.Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS microbiology reviews. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 2.Hood MI, Skaar EP. Nutritional immunity: transition metals at the pathogen-host interface. Nature reviews Microbiology. 2012;10:525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weinberg ED. Iron and susceptibility to infectious disease. Science. 1974;184:952–956. doi: 10.1126/science.184.4140.952. [DOI] [PubMed] [Google Scholar]
- 4.Dobryszycka W. Biological functions of haptoglobin–new pieces to an old puzzle. European journal of clinical chemistry and clinical biochemistry: journal of the Forum of European Clinical Chemistry Societies. 1997;35:647–654. [PubMed] [Google Scholar]
- 5.Tolosano E, Altruda F. Hemopexin: structure, function, and regulation. DNA and cell biology. 2002;21:297–306. doi: 10.1089/104454902753759717. [DOI] [PubMed] [Google Scholar]
- 6.Snow GA. Mycobactins: iron-chelating growth factors from mycobacteria. Bacteriological reviews. 1970;34:99–125. doi: 10.1128/br.34.2.99-125.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Madigan CA, Cheng TY, Layre E, Young DC, McConnell MJ, Debono CA, Murry JP, Wei JR, Barry CE, 3rd, Rodriguez GM, Matsunaga I, Rubin EJ, Moody DB. Lipidomic discovery of deoxysiderophores reveals a revised mycobactin biosynthesis pathway in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2012;109:1257–1262. doi: 10.1073/pnas.1109958109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McMahon MD, Rush JS, Thomas MG. Analyses of MbtB, MbtE, and MbtF suggest revisions to the mycobactin biosynthesis pathway in Mycobacterium tuberculosis. Journal of bacteriology. 2012;194:2809–2818. doi: 10.1128/JB.00088-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quadri LE, Sello J, Keating TA, Weinreb PH, Walsh CT. Identification of a Mycobacterium tuberculosis gene cluster encoding the biosynthetic enzymes for assembly of the virulence-conferring siderophore mycobactin. Chemistry & biology. 1998;5:631–645. doi: 10.1016/s1074-5521(98)90291-5. [DOI] [PubMed] [Google Scholar]
- 10.Gobin J, Moore CH, Reeve JR, Jr, Wong DK, Gibson BW, Horwitz MA. Iron acquisition by Mycobacterium tuberculosis: isolation and characterization of a family of iron-binding exochelins. Proc Natl Acad Sci U S A. 1995;92:5189–5193. doi: 10.1073/pnas.92.11.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ratledge C, Dover LG. Iron metabolism in pathogenic bacteria. Annual review of microbiology. 2000;54:881–941. doi: 10.1146/annurev.micro.54.1.881. [DOI] [PubMed] [Google Scholar]
- 12.Gobin J, Horwitz MA. Exochelins of Mycobacterium tuberculosis remove iron from human iron-binding proteins and donate iron to mycobactins in the M. tuberculosis cell wall. The Journal of experimental medicine. 1996;183:1527–1532. doi: 10.1084/jem.183.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodriguez GM, Gardner R, Kaur N, Phanstiel Ot. Utilization of Fe3+-acinetoferrin analogs as an iron source by Mycobacterium tuberculosis. Biometals: an international journal on the role of metal ions in biology, biochemistry, and medicine. 2008;21:93–103. doi: 10.1007/s10534-007-9096-5. [DOI] [PubMed] [Google Scholar]
- 14.Wells RM, Jones CM, Xi Z, Speer A, Danilchanka O, Doornbos KS, Sun P, Wu F, Tian C, Niederweis M. Discovery of a Siderophore Export System Essential for Virulence of Mycobacterium tuberculosis. PLoS pathogens. 2013;9:e1003120. doi: 10.1371/journal.ppat.1003120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Asou N, Sanada I, Tanaka K, Hidaka M, Suzushima H, Matsuzaki H, Kawano F, Takatsuki K. Inversion of chromosome 16 and bone marrow eosinophilia in a myelomonocytic transformation of chronic myeloid leukemia. Cancer genetics and cytogenetics. 1992;61:197–200. doi: 10.1016/0165-4608(92)90086-n. [DOI] [PubMed] [Google Scholar]
- 16.Ryndak MB, Wang S, Smith I, Rodriguez GM. The Mycobacterium tuberculosis high-affinity iron importer, IrtA, contains an FAD-binding domain. Journal of bacteriology. 2010;192:861–869. doi: 10.1128/JB.00223-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Serafini A, Boldrin F, Palu G, Manganelli R. Characterization of a Mycobacterium tuberculosis ESX-3 conditional mutant: essentiality and rescue by iron and zinc. Journal of bacteriology. 2009;191:6340–6344. doi: 10.1128/JB.00756-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siegrist MS, Unnikrishnan M, McConnell MJ, Borowsky M, Cheng TY, Siddiqi N, Fortune SM, Moody DB, Rubin EJ. Mycobacterial Esx-3 is required for mycobactin-mediated iron acquisition. Proc Natl Acad Sci U S A. 2009;106:18792–18797. doi: 10.1073/pnas.0900589106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones CM, Niederweis M. Mycobacterium tuberculosis can utilize heme as an iron source. Journal of bacteriology. 2011;193:1767–1770. doi: 10.1128/JB.01312-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tullius MV, Harmston CA, Owens CP, Chim N, Morse RP, McMath LM, Iniguez A, Kimmey JM, Sawaya MR, Whitelegge JP, Horwitz MA, Goulding CW. Discovery and characterization of a unique mycobacterial heme acquisition system. Proc Natl Acad Sci U S A. 2011;108:5051–5056. doi: 10.1073/pnas.1009516108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nambu S, Matsui T, Goulding CW, Takahashi S, Ikeda-Saito M. A new way to degrade heme: The Mycobacterium tuberculosis enzyme MhuD catalyzes heme degradation without generating CO. The Journal of biological chemistry. 2013 doi: 10.1074/jbc.M112.448399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiancone E, Ceci P, Ilari A, Ribacchi F, Stefanini S. Iron and proteins for iron storage and detoxification. Biometals: an international journal on the role of metal ions in biology, biochemistry, and medicine. 2004;17:197–202. doi: 10.1023/b:biom.0000027692.24395.76. [DOI] [PubMed] [Google Scholar]
- 23.Gupta V, Gupta RK, Khare G, Salunke DM, Tyagi AK. Crystal structure of Bfr A from Mycobacterium tuberculosis: incorporation of selenomethionine results in cleavage and demetallation of haem. PLoS One. 2009;4:e8028. doi: 10.1371/journal.pone.0008028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khare G, Gupta V, Nangpal P, Gupta RK, Sauter NK, Tyagi AK. Ferritin structure from Mycobacterium tuberculosis: comparative study with homologues identifies extended C-terminus involved in ferroxidase activity. PLoS One. 2011;6:e18570. doi: 10.1371/journal.pone.0018570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pandey R, Rodriguez GM. A ferritin mutant of Mycobacterium tuberculosis is highly susceptible to killing by antibiotics and is unable to establish a chronic infection in mice. Infection and immunity. 2012;80:3650–3659. doi: 10.1128/IAI.00229-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reddy PV, Puri RV, Khera A, Tyagi AK. Iron storage proteins are essential for the survival and pathogenesis of Mycobacterium tuberculosis in THP-1 macrophages and the guinea pig model of infection. Journal of bacteriology. 2012;194:567–575. doi: 10.1128/JB.05553-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takatsuka M, Osada-Oka M, Satoh EF, Kitadokoro K, Nishiuchi Y, Niki M, Inoue M, Iwai K, Arakawa T, Shimoji Y, Ogura H, Kobayashi K, Rambukkana A, Matsumoto S. A histone-like protein of mycobacteria possesses ferritin superfamily protein-like activity and protects against DNA damage by Fenton reaction. PLoS One. 2011;6:e20985. doi: 10.1371/journal.pone.0020985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez GM, Voskuil MI, Gold B, Schoolnik GK, Smith I. ideR, An essential gene in mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infection and immunity. 2002;70:3371–3381. doi: 10.1128/IAI.70.7.3371-3381.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmitt MP, Predich M, Doukhan L, Smith I, Holmes RK. Characterization of an iron-dependent regulatory protein (IdeR) of Mycobacterium tuberculosis as a functional homolog of the diphtheria toxin repressor (DtxR) from Corynebacterium diphtheriae. Infection and immunity. 1995;63:4284–4289. doi: 10.1128/iai.63.11.4284-4289.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pohl E, Holmes RK, Hol WG. Crystal structure of the iron-dependent regulator (IdeR) from Mycobacterium tuberculosis shows both metal binding sites fully occupied. J Mol Biol. 1999;285:1145–1156. doi: 10.1006/jmbi.1998.2339. [DOI] [PubMed] [Google Scholar]
- 31.Semavina M, Beckett D, Logan TM. Metal-linked dimerization in the iron-dependent regulator from Mycobacterium tuberculosis. Biochemistry. 2006;45:12480–12490. doi: 10.1021/bi060797s. [DOI] [PubMed] [Google Scholar]
- 32.Gold B, Rodriguez GM, Marras SA, Pentecost M, Smith I. The Mycobacterium tuberculosis IdeR is a dual functional regulator that controls transcription of genes involved in iron acquisition, iron storage and survival in macrophages. Molecular microbiology. 2001;42:851–865. doi: 10.1046/j.1365-2958.2001.02684.x. [DOI] [PubMed] [Google Scholar]
- 33.Petrera A, Amstutz B, Gioia M, Hahnlein J, Baici A, Selchow P, Ferraris DM, Rizzi M, Sbardella D, Marini S, Coletta M, Sander P. Functional characterization of the Mycobacterium tuberculosis zinc metallopeptidase Zmp1 and identification of potential substrates. Biological chemistry. 2012;393:631–640. doi: 10.1515/hsz-2012-0106. [DOI] [PubMed] [Google Scholar]
- 34.Srinivasan R, Anilkumar G, Rajeswari H, Ajitkumar P. Functional characterization of AAA family FtsH protease of Mycobacterium tuberculosis. FEMS Microbiol Lett. 2006;259:97–105. doi: 10.1111/j.1574-6968.2006.00251.x. [DOI] [PubMed] [Google Scholar]
- 35.Supuran CT. Carbonic anhydrases–an overview. Current pharmaceutical design. 2008;14:603–614. doi: 10.2174/138161208783877884. [DOI] [PubMed] [Google Scholar]
- 36.Pegan SD, Rukseree K, Franzblau SG, Mesecar AD. Structural basis for catalysis of a tetrameric class IIa fructose 1,6-bisphosphate aldolase from Mycobacterium tuberculosis. J Mol Biol. 2009;386:1038–1053. doi: 10.1016/j.jmb.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ordonez H, Unciuleac M, Shuman S. Mycobacterium smegmatis RqlH defines a novel clade of bacterial RecQ-like DNA helicases with ATP-dependent 3′–5′ translocase and duplex unwinding activities. Nucleic Acids Res. 2012;40:4604–4614. doi: 10.1093/nar/gks046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanchez-Quitian ZA, Schneider CZ, Ducati RG, de Azevedo WF, Jr, Bloch C, Jr, Basso LA, Santos DS. Structural and functional analyses of Mycobacterium tuberculosis Rv3315c-encoded metal-dependent homotetrameric cytidine deaminase. J Struct Biol. 2010;169:413–423. doi: 10.1016/j.jsb.2009.12.019. [DOI] [PubMed] [Google Scholar]
- 39.Tremblay LW, Fan F, Vetting MW, Blanchard JS. The 1.6 A crystal structure of Mycobacterium smegmatis MshC: the penultimate enzyme in the mycothiol biosynthetic pathway. Biochemistry. 2008;47:13326–13335. doi: 10.1021/bi801708f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buetow L, Brown AC, Parish T, Hunter WN. The structure of Mycobacteria 2C-methyl-D-erythritol-2,4-cyclodiphosphate synthase, an essential enzyme, provides a platform for drug discovery. BMC structural biology. 2007;7:68. doi: 10.1186/1472-6807-7-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koon N, Squire CJ, Baker EN. Crystal structure of LeuA from Mycobacterium tuberculosis, a key enzyme in leucine biosynthesis. Proc Natl Acad Sci U S A. 2004;101:8295–8300. doi: 10.1073/pnas.0400820101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Piddington DL, Fang FC, Laessig T, Cooper AM, Orme IM, Buchmeier NA. Cu,Zn superoxide dismutase of Mycobacterium tuberculosis contributes to survival in activated macrophages that are generating an oxidative burst. Infection and immunity. 2001;69:4980–4987. doi: 10.1128/IAI.69.8.4980-4987.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu CH, Tsai-Wu JJ, Huang YT, Lin CY, Lioua GG, Lee FJ. Identification and subcellular localization of a novel Cu,Zn superoxide dismutase of Mycobacterium tuberculosis. FEBS Lett. 1998;439:192–196. doi: 10.1016/s0014-5793(98)01373-8. [DOI] [PubMed] [Google Scholar]
- 44.Ilghari D, Lightbody KL, Veverka V, Waters LC, Muskett FW, Renshaw PS, Carr MD. Solution structure of the Mycobacterium tuberculosis EsxG.EsxH complex: functional implications and comparisons with other M. tuberculosis Esx family complexes. The Journal of biological chemistry. 2011;286:29993–30002. doi: 10.1074/jbc.M111.248732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Norman RA, McAlister MS, Murray-Rust J, Movahedzadeh F, Stoker NG, McDonald NQ. Crystal structure of inositol 1-phosphate synthase from Mycobacterium tuberculosis, a key enzyme in phosphatidylinositol synthesis. Structure. 2002;10:393–402. doi: 10.1016/s0969-2126(02)00718-9. [DOI] [PubMed] [Google Scholar]
- 46.Zhang L, Xiao N, Pan Y, Zheng Y, Pan Z, Luo Z, Xu X, Liu Y. Binding and inhibition of copper ions to RecA inteins from Mycobacterium tuberculosis. Chemistry. 2010;16:4297–4306. doi: 10.1002/chem.200903584. [DOI] [PubMed] [Google Scholar]
- 47.Hantke K. Bacterial zinc uptake and regulators. Current opinion in microbiology. 2005;8:196–202. doi: 10.1016/j.mib.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 48.Nies DH. Efflux-mediated heavy metal resistance in prokaryotes. FEMS microbiology reviews. 2003;27:313–339. doi: 10.1016/S0168-6445(03)00048-2. [DOI] [PubMed] [Google Scholar]
- 49.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 50.Verkhovtseva NV, Filina N, Pukhov DE. Evolutionary role of iron in metabolism of prokaryotes and biogeochemical processes. Zhurnal evoliutsionnoi biokhimii i fiziologii. 2001;37:338–343. [PubMed] [Google Scholar]
- 51.Cavet JS, Meng W, Pennella MA, Appelhoff RJ, Giedroc DP, Robinson NJ. A nickel-cobalt-sensing ArsR-SmtB family repressor. Contributions of cytosol and effector binding sites to metal selectivity. The Journal of biological chemistry. 2002;277:38441–38448. doi: 10.1074/jbc.M207677200. [DOI] [PubMed] [Google Scholar]
- 52.Botella H, Peyron P, Levillain F, Poincloux R, Poquet Y, Brandli I, Wang C, Tailleux L, Tilleul S, Charriere GM, Waddell SJ, Foti M, Lugo-Villarino G, Gao Q, Maridonneau-Parini I, Butcher PD, Castagnoli PR, Gicquel B, de Chastellier C, Neyrolles O. Mycobacterial p(1)-type ATPases mediate resistance to zinc poisoning in human macrophages. Cell host & microbe. 2011;10:248–259. doi: 10.1016/j.chom.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ward SK, Abomoelak B, Hoye EA, Steinberg H, Talaat AM. CtpV: a putative copper exporter required for full virulence of Mycobacterium tuberculosis. Molecular microbiology. 2010;77:1096–1110. doi: 10.1111/j.1365-2958.2010.07273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fu Y, Tsui HC, Bruce KE, Sham LT, Higgins KA, Lisher JP, Kazmierczak KM, Maroney MJ, Dann CE, 3rd, Winkler ME, Giedroc DP. A new structural paradigm in copper resistance in Streptococcus pneumoniae. Nature chemical biology. 2013;9:177–183. doi: 10.1038/nchembio.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Festa RA, Jones MB, Butler-Wu S, Sinsimer D, Gerads R, Bishai WR, Peterson SN, Darwin KH. A novel copper-responsive regulon in Mycobacterium tuberculosis. Molecular microbiology. 2011;79:133–148. doi: 10.1111/j.1365-2958.2010.07431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ward SK, Hoye EA, Talaat AM. The global responses of Mycobacterium tuberculosis to physiological levels of copper. Journal of bacteriology. 2008;190:2939–2946. doi: 10.1128/JB.01847-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu T, Ramesh A, Ma Z, Ward SK, Zhang L, George GN, Talaat AM, Sacchettini JC, Giedroc DP. CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nature chemical biology. 2007;3:60–68. doi: 10.1038/nchembio844. [DOI] [PubMed] [Google Scholar]
- 58.Wolschendorf F, Ackart D, Shrestha TB, Hascall-Dove L, Nolan S, Lamichhane G, Wang Y, Bossmann SH, Basaraba RJ, Niederweis M. Copper resistance is essential for virulence of Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2011;108:1621–1626. doi: 10.1073/pnas.1009261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Botella H, Stadthagen G, Lugo-Villarino G, de Chastellier C, Neyrolles O. Metallobiology of host-pathogen interactions: an intoxicating new insight. Trends in microbiology. 2012;20:106–112. doi: 10.1016/j.tim.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 60.Rowland JL, Niederweis M. Resistance mechanisms of Mycobacterium tuberculosis against phagosomal copper overload. Tuberculosis (Edinb) 2012;92:202–210. doi: 10.1016/j.tube.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Samanovic MI, Ding C, Thiele DJ, Darwin KH. Copper in microbial pathogenesis: meddling with the metal. Cell host & microbe. 2012;11:106–115. doi: 10.1016/j.chom.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.White C, Lee J, Kambe T, Fritsche K, Petris MJ. A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity. The Journal of biological chemistry. 2009;284:33949–33956. doi: 10.1074/jbc.M109.070201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Padilla-Benavides T, Long JE, Raimunda D, Sassetti CM, Arguello JM. A novel P1B-type Mn2+ transporting ATPase is required for secreted protein metallation in mycobacteria. The Journal of biological chemistry. 2013 doi: 10.1074/jbc.M112.448175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Francis MS, Thomas CJ. Mutants in the CtpA copper transporting P-type ATPase reduce virulence of Listeria monocytogenes. Microbial pathogenesis. 1997;22:67–78. doi: 10.1006/mpat.1996.0092. [DOI] [PubMed] [Google Scholar]
- 65.Schwan WR, Warrener P, Keunz E, Stover CK, Folger KR. Mutations in the cueA gene encoding a copper homeostasis P-type ATPase reduce the pathogenicity of Pseudomonas aeruginosa in mice. International journal of medical microbiology: IJMM. 2005;295:237–242. doi: 10.1016/j.ijmm.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 66.Shafeeq S, Yesilkaya H, Kloosterman TG, Narayanan G, Wandel M, Andrew PW, Kuipers OP, Morrissey JA. The cop operon is required for copper homeostasis and contributes to virulence in Streptococcus pneumoniae. Molecular microbiology. 2011;81:1255–1270. doi: 10.1111/j.1365-2958.2011.07758.x. [DOI] [PubMed] [Google Scholar]
- 67.Osman D, Waldron KJ, Denton H, Taylor CM, Grant AJ, Mastroeni P, Robinson NJ, Cavet JS. Copper homeostasis in Salmonella is atypical and copper-CueP is a major periplasmic metal complex. The Journal of biological chemistry. 2010;285:25259–25268. doi: 10.1074/jbc.M110.145953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Macomber L, Rensing C, Imlay JA. Intracellular copper does not catalyze the formation of oxidative DNA damage in Escherichia coli. Journal of bacteriology. 2007;189:1616–1626. doi: 10.1128/JB.01357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chillappagari S, Seubert A, Trip H, Kuipers OP, Marahiel MA, Miethke M. Copper stress affects iron homeostasis by destabilizing iron-sulfur cluster formation in Bacillus subtilis. Journal of bacteriology. 2010;192:2512–2524. doi: 10.1128/JB.00058-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu FF, Imlay JA. Silver(I), mercury(II), cadmium(II), and zinc(II) target exposed enzymic iron-sulfur clusters when they toxify Escherichia coli. Applied and environmental microbiology. 2012;78:3614–3621. doi: 10.1128/AEM.07368-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McDevitt CA, Ogunniyi AD, Valkov E, Lawrence MC, Kobe B, McEwan AG, Paton JC. A molecular mechanism for bacterial susceptibility to zinc. PLoS pathogens. 2011;7:e1002357. doi: 10.1371/journal.ppat.1002357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Osman D, Patterson CJ, Bailey K, Fisher K, Robinson NJ, Rigby SE, Cavet JS. The copper supply pathway to a Salmonella Cu,Zn-superoxide dismutase (SodCII) involves P(1B)-type ATPase copper efflux and periplasmic CueP. Molecular microbiology. 2013;87:466–477. doi: 10.1111/mmi.12107. [DOI] [PubMed] [Google Scholar]
- 73.Lucarelli D, Russo S, Garman E, Milano A, Meyer-Klaucke W, Pohl E. Crystal structure and function of the zinc uptake regulator FurB from Mycobacterium tuberculosis. The Journal of biological chemistry. 2007;282:9914–9922. doi: 10.1074/jbc.M609974200. [DOI] [PubMed] [Google Scholar]
- 74.Maciag A, Dainese E, Rodriguez GM, Milano A, Provvedi R, Pasca MR, Smith I, Palu G, Riccardi G, Manganelli R. Global analysis of the Mycobacterium tuberculosis Zur (FurB) regulon. Journal of bacteriology. 2007;189:730–740. doi: 10.1128/JB.01190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Milano A, Branzoni M, Canneva F, Profumo A, Riccardi G. The Mycobacterium tuberculosis Rv2358-furB operon is induced by zinc. Research in microbiology. 2004;155:192–200. doi: 10.1016/j.resmic.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 76.Pym AS, Domenech P, Honore N, Song J, Deretic V, Cole ST. Regulation of catalase-peroxidase (KatG) expression, isoniazid sensitivity and virulence by furA of Mycobacterium tuberculosis. Molecular microbiology. 2001;40:879–889. doi: 10.1046/j.1365-2958.2001.02427.x. [DOI] [PubMed] [Google Scholar]
- 77.Sala C, Forti F, Di Florio E, Canneva F, Milano A, Riccardi G, Ghisotti D. Mycobacterium tuberculosis FurA autoregulates its own expression. Journal of bacteriology. 2003;185:5357–5362. doi: 10.1128/JB.185.18.5357-5362.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zahrt TC, Song J, Siple J, Deretic V. Mycobacterial FurA is a negative regulator of catalase-peroxidase gene katG. Molecular microbiology. 2001;39:1174–1185. doi: 10.1111/j.1365-2958.2001.02321.x. [DOI] [PubMed] [Google Scholar]
- 79.Massonet C, Pintens V, Merckx R, Anne J, Lammertyn E, Van Eldere J. Effect of iron on the expression of sirR and sitABC in biofilm-associated Staphylococcus epidermidis. BMC microbiology. 2006;6:103. doi: 10.1186/1471-2180-6-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Campbell DR, Chapman KE, Waldron KJ, Tottey S, Kendall S, Cavallaro G, Andreini C, Hinds J, Stoker NG, Robinson NJ, Cavet JS. Mycobacterial cells have dual nickel-cobalt sensors: sequence relationships and metal sites of metal-responsive repressors are not congruent. The Journal of biological chemistry. 2007;282:32298–32310. doi: 10.1074/jbc.M703451200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cavet JS, Graham AI, Meng W, Robinson NJ. A cadmium-lead-sensing ArsR-SmtB repressor with novel sensory sites. Complementary metal discrimination by NmtR AND CmtR in a common cytosol. The Journal of biological chemistry. 2003;278:44560–44566. doi: 10.1074/jbc.M307877200. [DOI] [PubMed] [Google Scholar]
- 82.Canneva F, Branzoni M, Riccardi G, Provvedi R, Milano A. Rv2358 and FurB: two transcriptional regulators from Mycobacterium tuberculosis which respond to zinc. Journal of bacteriology. 2005;187:5837–5840. doi: 10.1128/JB.187.16.5837-5840.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wagner D, Maser J, Lai B, Cai Z, Barry CE, 3rd, Honer Zu Bentrup K, Russell DG, Bermudez LE. Elemental analysis of Mycobacterium avium-, Mycobacterium tuberculosis-, and Mycobacterium smegmatis-containing phagosomes indicates pathogen-induced microenvironments within the host cell’s endosomal system. Journal of immunology. 2005;174:1491–1500. doi: 10.4049/jimmunol.174.3.1491. [DOI] [PubMed] [Google Scholar]
- 84.Raimunda D, Long JE, Sassetti CM, Arguello JM. Role in metal homeostasis of CtpD, a Co(2)(+) transporting P(1B4)-ATPase of Mycobacterium smegmatis. Molecular microbiology. 2012;84:1139–1149. doi: 10.1111/j.1365-2958.2012.08082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


