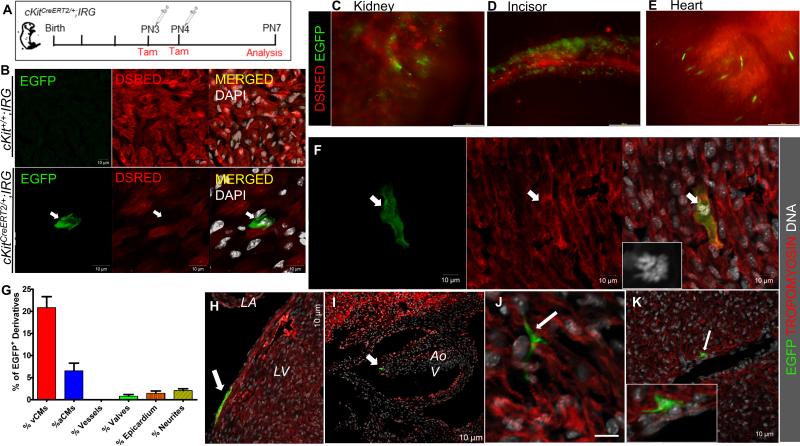
Figure 7. Genetic fate-map of postnatal cKit-expressing cells in the neonatal mouse.
A, Schematic of the genetic fate-mapping strategy. cKitCreERT2/+;IRG neonates (n=6) were pulsed with tamoxifen at PN3 and PN4 and expression of EGFP in cKit+ cells and their derivatives was analyzed on PN7. B. Representative confocal immunofluorescence of EGFP and DSRED in tamoxifen-treated cKit+/+;IRG (top) and cKitCreERT2/+;IRG (bottom) littermates. EGFP is exclusively expressed upon Cre-mediated recombination. C-E, Expression of EGFP was detected in all tissues in which contribution from cKit-expressing cells has been previously reported, including the kidney (C), the developing incisors (D) and the heart (E). F, confocal immunofluorescence of an EGFP+/Tropomyosin+ ventricular cardiomyocyte (arrow) in mitosis (inset, DNA). G, Quantification of EGFP+ derivatives in the postnatal heart (n=2). H-J, In addition to cardiomyocytes, EGFP+ derivatives of postnatal CSCs were also detected in the epicardium (H, arrow), cardiac valves (I, arrow) and in neuronal cardiac cells (J). K, Although EGFP cells were occasionally detected in close proximity to large coronary vessels (arrow), contribution of CSCs to coronary vascular cells was never detected. vCMs, ventricular cardiomyocytes; aCMs, atrial cardiomyocytes; LA, left atrium; LV, left ventricle; Aov, aortic valve. Data are presented as mean±SEM. Scale bars, 200μm (C-E); 10μm (B, F-K).