Abstract
Mathematical models, such as those that forecast the spread of epidemics or predict the weather, must overcome the challenges of integrating incomplete and inaccurate data in computer simulations, estimating the probability of multiple possible scenarios, incorporating changes in human behavior and/or the pathogen, and environmental factors. In the past 3 decades, the weather forecasting community has made significant advances in data collection, assimilating heterogeneous data steams into models and communicating the uncertainty of their predictions to the general public. Epidemic modelers are struggling with these same issues in forecasting the spread of emerging diseases, such as Zika virus infection and Ebola virus disease. While weather models rely on physical systems, data from satellites, and weather stations, epidemic models rely on human interactions, multiple data sources such as clinical surveillance and Internet data, and environmental or biological factors that can change the pathogen dynamics. We describe some of similarities and differences between these 2 fields and how the epidemic modeling community is rising to the challenges posed by forecasting to help anticipate and guide the mitigation of epidemics. We conclude that some of the fundamental differences between these 2 fields, such as human behavior, make disease forecasting more challenging than weather forecasting.
Keywords: disease, weather, forecasting, Internet data, modeling
Infectious diseases have plagued humankind for millennia, but we have yet to understand many of the factors driving their transmission and predicting their course. The recent emergence of Ebola, chikungunya, and Zika viruses highlight the importance of quickly identifying outbreaks of infection and mitigating their impact. Internet data streams (IDS), such as Google search volumes and Twitter feeds, can help epidemic models derive better predictions to perhaps ultimately forecast the future of an outbreak and guide mitigation strategies.
Forecasting is the ability to predict what will happen in the future on the basis of analysis of past and current data. Weather forecasting has improved tremendously in the past 3 decades, supported by billions of dollars of investment in data collection and advances in numerical algorithms, software, and computer power [1, 2]. Today, weather forecasts provide useful answers to questions such as “Will it rain tomorrow?” or “How much snow will fall this winter?”
Epidemic models have historically been used to understand the spread and control of diseases [3]. Recently, they are being applied to provide answers to predictive questions such as “How many people will be sick with the flu next week?” and “Will the outbreak peak next month?” This type of information can help public health officials manage limited staff and resources during the influenza season.
Researchers are borrowing approaches used for weather forecasting to improve epidemic models and quantify the uncertainty in these predictions [4]. The promising results from these approaches have led policy makers and the epidemic modeling community to draw comparisons between real-time weather and disease forecasting [5].
Both the weather and disease incidence can be forecasted using nonlinear dynamics and informed by real-time observations. Weather forecasting models assimilate a continuous stream of data from weather stations. Similarly, epidemic models can dynamically incorporate a continuous stream of data (eg, clinical surveillance and IDS) to model and forecast disease incidence. The established practices in data assimilation and uncertainty quantification for weather modeling have the potential to guide scientists in using IDS in the next generation of epidemic models.
There are major differences between weather and epidemic models. Weather models are based on physical principles for particles, which follow a prescribed set of nonlinear dynamical laws. The transmission dynamics in epidemic models are governed by human social behavior, which can change during the course of an epidemic. These behavior changes can have an immediate impact on the future of an epidemic. Thus, to accurately forecast an emerging epidemic, we need to forecast the behavior of individuals, potential changes in the pathogen, and their interactions as they relate to the transmission dynamics.
A major difference between weather and disease forecasting is the availability and quality of ground truth data. Clinical surveillance data (used for calibration and verification of epidemic models) are much less accurate than the atmospheric data available for weather models. Typically, the data available in the early stages of an emerging epidemic are for the number of new cases. This type of information is insufficient to accurately calibrate all the parameters in a disease transmission model, such the susceptible and exposed populations.
Another difference between these 2 domains is that the public is accustomed to hearing and understanding probabilistic weather forecasts, such as “there is a 20% chance of rain this afternoon.” Epidemic forecasts do not have a similar foundation to communicate disease forecasts and, most importantly, the uncertainty in the predictions to decision makers or the public at large. The use of spatial risk maps for the probability of an outbreak and the severity of the potential outbreak, as well as so-called what-if-scenario–type approaches, are beginning to be used in new and innovative ways to fill this gap, but they are still a long way from being as well understood and useful as the current weather maps and evening weather forecasts.
Weather Forecasting
Weather forecasting technology has advanced so that the forecasts are easy to understand and ubiquitous in newspapers, on television, websites, smartphone apps, and even airplane boarding passes. Among US smartphone users, the most commonly accessed digital content is weather information [6]. Simply typing “weather” in the Google search engine creates a user-friendly display of the current local weather, complete with hourly and daily forecasts. There is not an analogous capability to find out the local incidence or risk of being infected by all diseases around the globe.
While the general public sees only the simplified output of weather forecasts, the underpinnings of these storm cloud pictures and temperature plots are based on complex nonlinear computer models built by teams of scientists over decades. Although computer models have improved weather forecasting accuracy, long-term and even annual and seasonal forecasts remain a challenge. Short-term predictions of up to 2 weeks are generally regarded as the limit for accurate local-level weather predictions, with uncertainty growing the further out a forecast goes [7].
The process for weather forecasting can be broken into 5 steps: (1) observations, (2) analysis of those observations, (3) models, (4) forecasts, and (5) updates. Observations for weather forecasting are gathered from thousands of automated weather stations on land, at sea, and from satellites collecting information from the atmosphere, ocean, land, and cryosphere [8]. These data are freely available for anyone to use. The World Meteorological Organization standardizes the instrumentation, observing practices, and timing of these observations [9]. Because weather observation can largely be automated, sensors can record data at high resolution and provide worldwide hourly weather data for public consumption [10]. Meteorologists have created tools for dealing with the masses of data produced by all of these observations. For example, on a daily basis, scientists at the National Oceanic and Atmospheric Administration sift through mountains of data from thousands of sensors all over the world.
There are vetted mathematical models designed to use observations for weather prediction, including the UKMET Unified Model, the European Center for Medium-Range Weather Forecasts model, and the Global Forecast Systems model. These models are all based on a set of primitive equations that are used to predict the physics and dynamics of the atmosphere [8].
Forecasts commonly take the form of an ensemble (a collection of possible outcomes) of predictions. These forecasts are often evaluated using probabilistic forecasting metrics, such as their Brier or relative operating characteristic scores [11] for dichotomous events (eg, “Will the temperature exceed some threshold by next week?”) or probability estimates for continuous events [12] (eg, “What will the temperature be tomorrow?”). Validating forecasts from multiple models with a broad range of metrics allows the forecasters to identify and address their model's deficiencies. The high volume of forecasts provides rapid feedback on the quality of the predictions and guides the forecasters in calibrating and improving their models.
The impact of human behavior on weather forecasts is only visible over long periods. For example, carrying an umbrella today will not affect the likelihood that rain will fall tomorrow, but human behaviors affecting atmospheric pollution typically influence long-range weather patterns. Therefore, the changes caused by human factors are generally not considered in short- and medium-range weather forecasts.
Epidemic Forecasting
Disease models are also created as systems of complex nonlinear equations. The epidemic modeling community is less mature than the current weather modeling community; epidemic models are probably close to the state that weather models were in the early 1970s. The transmission model parameters are typically estimated using official clinical surveillance data. Disease surveillance systems rely on various sources, including patient interviews, sentinel medical provider reports, and laboratory tests. Resulting data are sent up a bureaucratic reporting chain, first reaching the desks of public health officials and government employees. Data are available to clinicians, researchers, and the general public days, weeks, or even months after initial recording [13]. These data are generally aggregated at various geographic resolutions, owing to privacy concerns. This process, although generally considered accurate once complete, is expensive, leads to a significant lag between observation and reporting, and prevents researchers from providing optimal decision support due to the aggregated nature of the data.
The shortcomings of disease surveillance systems have led to the adoption of IDS, such as search queries and social media posts, for epidemic forecasting [14–17]. Whereas official data streams are generally expensive and unavailable in real time, IDS are often free or inexpensive and available in real time. This difference can be partly attributed to the fact that the user automatically drives the information provided by health-related IDS, whereas medical or laboratory workers collect official disease information.
The use of IDS in disease surveillance is a new and growing field. Myriad studies within the past decade adopted the concept of identifying and extracting health-related activity traces in Internet data for the purpose of disease monitoring and forecasting [17]. Most of the research in epidemic forecasting using IDS has been done using Google search queries [14, 18] and Twitter posts [15, 19], but Wikipedia [16, 20], other search engines, such as Baidu [21], and prediction markets [22] have also been explored as sources for Internet-based user-driven data. One limiting factor to using sources such as blogs, review websites, crowdsourcing applications, and other social media sites is the ability to gather, process, and understand the vast amount of data.
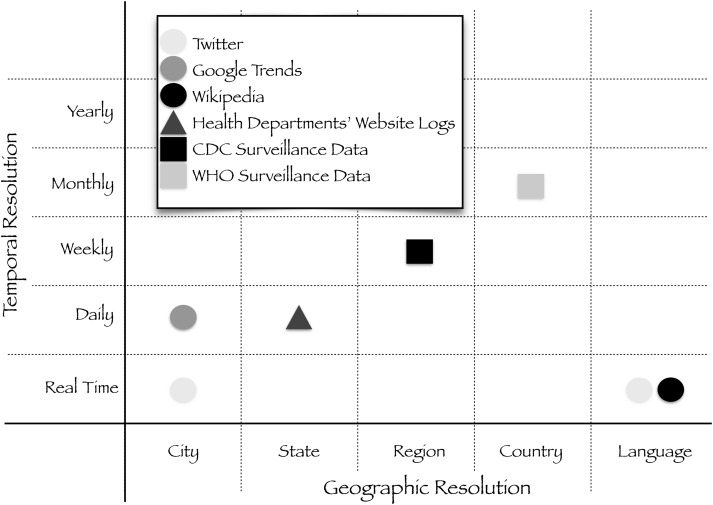
Today, IDS are far less reliable than the data collected from weather stations, owing to the lack of standardization and geographic information, bias, and inability to verify or clarify the information provided (Figure 1). For example, Wikipedia access logs are not currently geolocated, so language must be used as a proxy for location when using Wikipedia data. Similarly, a very small fraction of tweets are geolocated. There are biases associated with each IDS, from age, sex, and race to social status and global reach [23].
Figure 1.
Schematic representation of data streams used for epidemic modeling. Data streams vary by geographic and temporal resolution, as well as demographic differentiation and global reach. The epidemic modeling community would benefit from having real-time clinical surveillance data at the city or hospital level. Abbrevaitions: CDC, Centers for Disease Control and Prevention; WHO, World Health Organization.
In addition, language and cultural differences are barriers to finding the right information. The simplest approach for extracting information from unstructured data, such as tweets, is the bag-of-words approach, in which the frequencies of certain words (or posts containing said words) are tallied. The downfall to this approach is its inability to infer context; the bag-of-words approach cannot tell “That guy on the bus coughed all over me, and now I have a fever” from “That concert gave me raging Bieber fever” [24]. References to Bieber fever provide no useful information for forecasting levels of influenza-like illness in the United States. Other, more-advanced natural language–processing algorithms for tweet classification are capable of inferring context, recognizing events, and deducing sentiments and opinions.
Internet-based disease models are sensitive to events that lead to unusual behavior, such as pandemics or other events of high media interest. Such events can derail these models if they are not subject to frequent retraining and used under careful scrutiny for changes in algorithm dynamics [17]. The failure of Google Flu Trends during the 2009 influenza A(H1N1) pandemic is an example of this phenomenon [25].
Long-term disease forecasts can predict the peak of the coming year's influenza season or its severity [4]. Short-term forecasts can be used by hospitals to anticipate the case burden in the coming week. In disease forecasting, “nowcasting” refers to predicting current disease levels in absence of official data. IDS add value to epidemic forecasting models because they often provide information before official data are released. Furthermore, IDS have the potential to add finer levels of geographic and temporal resolution to epidemic forecasts.
National-level season long forecasts are of interest to policy makers deciding how much funding to allocate for pandemic vaccine production. On the other hand, city-level daily forecasts can be useful to hospitals making daily staffing decisions. An advantage of IDS is that it is often available in near real time for very local regions, while official disease incidence data may not be available at the ideal geographic or temporal granularity needed to address an emerging epidemic. However, it is worth noting that the availability of IDS varies substantially by geographic region, owing to limited Internet penetration in many developing countries.
The process for epidemic forecasting requires the same general steps as weather forecasting. Disease observations are gathered from both traditional (eg, healthcare provided records and laboratory records) and nontraditional (eg, Internet search queries and social media) sources. These sensors have vastly different levels of uncertainty, specificity, resolution (both temporal and geographical), and credibility. Furthermore, there are far fewer observational data points for diseases than for weather. Many countries do not record or report disease surveillance data. Of the data that are reported, there are not international standards of observation and reporting. For example, many nations have different notifiable diseases, report these diseases in diverse formats, and define the epidemiological week differently [26].
No validated, epidemic forecasting production codes exist at even moderate scale, let alone at the extreme scale used daily for weather forecasting. The difference between the codes currently used for epidemic forecasting and weather forecasting is that they are research codes used for testing science questions, not production codes used for real-world forecasts. While supercomputers have been used for large agent-based models and to process and mine health-related IDS to incorporate in forecasts, there is no continuously dedicated platform for global disease forecasting. Since big data are relatively new to epidemiologists, this is unsurprising. However, the paradigm of epidemic forecasting does not necessarily need to involve centralized supercomputers. Perhaps a more appropriate approach, at least initially, is decentralized, such as one involving smaller models being run locally on desktops as needed by local health officials.
While mathematical and computational models are used to describe both disease and weather dynamics, there are agreed-upon physical models used to build weather forecasts. There is no consensus in the epidemic modeling community when it comes to choosing the most appropriate epidemic forecasting model, although compartmental SIR-type models and similar approaches [27] have been used to model disease dynamics. Similarly, there is no consensus on the best way to extract information from IDS. IDS-based epidemic forecasts have used myriad mathematical and statistical models to generate output, ranging from linear [18] and nonlinear [23] regression models to network-based predictions [28].
Although human actions over many years can influence the overall dynamics of weather, changes in human behavior have the potential to rapidly alter the course of an epidemic, skewing the forecasts. Studies have shown that risk perception can play a role in changing people's opinions about preventive measures to avoid infection, such as vaccination, face mask use, and hand washing [29]. These actions can in turn change transmission paths by reducing or eliminating the probability of infection [30]. For example, worldwide vaccination programs against smallpox entirely eliminated variola virus from circulation. Furthermore, changes in human behavior can be reflected in online information seeking and sharing. While Internet users may not generally search for influenza symptoms without being ill, the emergence of pandemic influenza could lead users to search for influenza symptoms out of fear or interest. Thus, human behavior shapes epidemic forecasts on 2 levels: by altering the course of the epidemic itself and by changing the nature of the IDS-based model inputs.
DISCUSSION
Epidemic forecasting is still in its infancy and is a growing field with great potential. The challenges for accurate epidemic forecasting include data availability and emergent changes in human behavior and pathogens. As shown in Figure 1, there are several data streams available that can inform models, but each of these data streams comes with many limitations. Capturing the uncertainties associated with each data stream is crucial for accurately fusing these data streams into models to derive forecasts. Our hope is that, as electronic health records and other types of data become more ubiquitous and available to each individual via their smart phones, there will be a parallel world, similar to that for weather forecasting, where billions of sensors will be uploading real-time information to obtain personalized disease forecasts. We conclude that epidemic forecasting is more challenging than weather forecasting, owing to the human component.
Notes
Acknowledgments. This article has been approved by Los Alamos National Laboratory for public release (LA-UR-16-23668).
Financial support. This work was supported by the Models of Infectious Disease Agent Study, National Institute of General Medical Sciences, National Institutes of Health (grant U01-GM097658-01). Los Alamos National Laboratory is operated by Los Alamos National Security, for the Department of Energy, under contract DE-AC52-06NA25396.
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.NCEP operational forecast skill. http://www.nco.ncep.noaa.gov/sib/verification/s1_scores/s1_scores.pdf Accessed 18 May 2016.
- 2.National Oceanographic and Atmospheric Administration. FY 2015 budget summary. http://goo.gl/TjHZnx Accessed 18 May 2016.
- 3.Germann TC, Kadau K, Longini IM, Macken CA. Mitigation strategies for pandemic influenza in the United States. Proc Natl Acad Sci U S A 2006; 103:5935–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaman J, Karspeck A. Forecasting seasonal outbreaks of influenza. Proc Natl Acad Sci U S A 2012; 109:20425–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.George D. Back to the future: Using historical dengue data to predict the next epidemic. 5 June 2015. https://goo.gl/1OXqq6 Accessed 18 May 2016.
- 6.Digital Content Next 2012. OPA study defines today's smartphone user. 20 August 2012. https://goo.gl/wX5UNl. Accessed 18 May 2016.
- 7.Dole R. Future forecasts, 2009. http://www.ncdc.noaa.gov/paleo/ctl/future2.html Accessed 8 March 2016.
- 8.Lynch P. The origins of computer weather prediction and climate modeling. J Comput Phys 2008; 227:3431–44. [Google Scholar]
- 9.World Meteorological Association. Standards (technical regulations). https://goo.gl/RYsk7M Accessed 18 May 2016.
- 10.Lott JN, Baldwin R. The FCC Integrated Surface Hourly Database, a new resource of global climate data In: 13th Symposium on Global Change and Climate Variations, Orlando, Florida, 2002. [Google Scholar]
- 11.Hamill TM, Juras J. Measuring forecast skill: is it real skill or is it the varying climatology? Q J R Meteorol Soc 2006; 132:2905–24. [Google Scholar]
- 12.Gneiting T, Balabdaoui F, Raftery AE. Probabilistic forecasts, calibration and sharpness. J R Stat Soc Series B Stat Methodol 2007; 69:243–68. [Google Scholar]
- 13.Jajosky RA, Groseclose SL. Evaluation of reporting timeliness of public health surveillance systems for infectious diseases. BMC Public Health 2004; 4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ginsberg J, Mohebbi MH, Patel RS, Brammer L, Smolinski MS, Brilliant L. Detecting influenza epidemics using search engine query data. Nature 2009; 457:1012–4. [DOI] [PubMed] [Google Scholar]
- 15.Salathe M, Khandelwal S. Assessing vaccination sentiments with online social media: implications for infectious disease dynamics and control. PLoS Comput Biol 2011; 7:e1002199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Generous N, Fairchild G, Deshpande A, Del Valle SY, Priedhorsky R. Global disease monitoring and forecasting with Wikipedia. PLoS Comput Biol 2014; 10:e1003892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Althouse B, Scarpino SV, Meyers LCA et al. . Enhancing disease surveillance with novel data streams: challenges and opportunities. EPJ Data Science 2015; 4:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Araz OM, Bentley D, Muelleman RL. Using google trends data in forecasting influenza-like-illness related visits in Omaha, Nebraska. Am J Emerg Med 2014; 32:1016–23. [DOI] [PubMed] [Google Scholar]
- 19.Signorini A, Segre AM, Polgreen PM. The use of twitter to track levels of disease activity and public concern in the U.S. during the influenza a H1N1 pandemic. PLoS One 2011; 6:e19467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McIver DJ, Brownstein JS. Wikipedia usage estimates prevalence of influenza-Like illness in the United States in near real-Time. PLoS Computat Biol 2014; 10:e1003581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jia-xing B, Ben-fu L, Geng P, Na L. Gonorrhea incidence forecasting research based on Baidu search data In: 2013 International Conference on Management Science and Engineering, Harbin, China, 17–19 July 2013. [Google Scholar]
- 22.Polgreen PM, Nelson FD, Neumann GR. Using prediction markets to forecast trends in infectious diseases. Microbe 2010; 1:459–65. [DOI] [PubMed] [Google Scholar]
- 23.Smith B, Smith TC, Gray GC, Ryan MAK. for the Millennium Cohort Study Team. When epidemiology meets the internet: web-based surveys in the millennium cohort study. Am J Epidemiol 2007; 166:1345–54. [DOI] [PubMed] [Google Scholar]
- 24.Culotta A. Lightweight methods to estimate influenza rates and alcohol sales volume from twitter messages. Lang Resour Eval 2013; 47:217–38. [Google Scholar]
- 25.Lazer D, Kennedy R, King G, Vespignani A. The parable of Google flu: Traps in big data analysis. Science 2014; 343:1203–5. [DOI] [PubMed] [Google Scholar]
- 26.Health Organization (WHO). WHO report on global surveillance of epidemic prone infectious diseases—introduction. http://www.who.int/csr/resources/publications/introduction/en/index4.html Accessed 8 March 2016.
- 27.Hethcote HW. The mathematics of infectious diseases. SIAM Review 2000; 42:599–653. [Google Scholar]
- 28.Davidson MW, Haim DA, Radin JM. Using networks to combine big data and traditional surveillance to improve influenza predictions. Nature 2015; 5:8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bish A, Michie S. Demographic and attitudinal determinants of protective behaviours during a pandemic: a review. Br J Health Psychol 2010; 15:797–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Del Valle S, Hethcote H, Hyman J, Castillo-Chavez C. Effects of behavioral changes in a smallpox attack model. Math Biosci 2005; 195:228–51. [DOI] [PubMed] [Google Scholar]