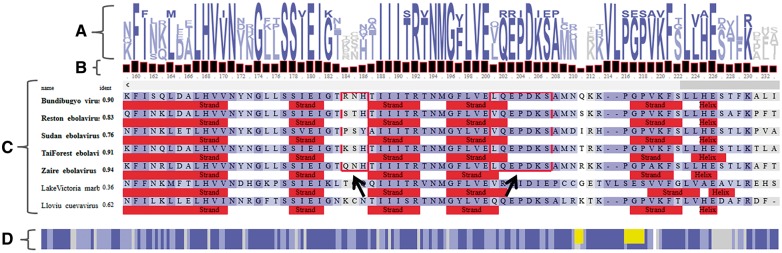
Fig. 1.
A simplified view of the MSAViewer for the sequence alignment of protein VP24 within seven viruses of Filoviridae family. (A) Sequence logo (Schneider and Stephens, 1990) representation with conservation patterns at each position in the MSA. (B) Bar chart showing amino acid conservation per position. (C) Main MSA panel with residues in the alignment colored according to the Percentage Identity coloring scheme (Waterhouse et al., 2009). Percentage sequence identities relative to consensus (consensus sequence not shown) are listed for each sequence. Filled red rectangles indicate sequence annotations provided by the user [here: secondary structure predictions of PredictProtein (Yachdav et al., 2014)]. Red frames (indicated by two black arrows) are clusters of residues of ebolavirus VP24 responsible for binding to karyopherin alpha nuclear transporters to suppress antiviral defense mechanism in human (Xu et al., 2014). (D) A compact overview MSA showing a bird’s eye view of the full alignment. Yellow rectangles are two highlighted clusters in the main MSA panel. Despite the overall sequence homology, the highlighted regions in ebolavirus (EBOV) proteins binding karyoprotein differ from those in Llovu cuevavirus (LLOV) and Lake Victoria marburgvirus (MARV). MARV is known to not suppress host’s antiviral defense mechanism (Xu et al., 2014). Though no experimental information on binding of LLOV VP24 to karyoproteins is available to date, the comparison of its sequence and structural features suggests a mechanism that is more similar to EBOV than to MARV