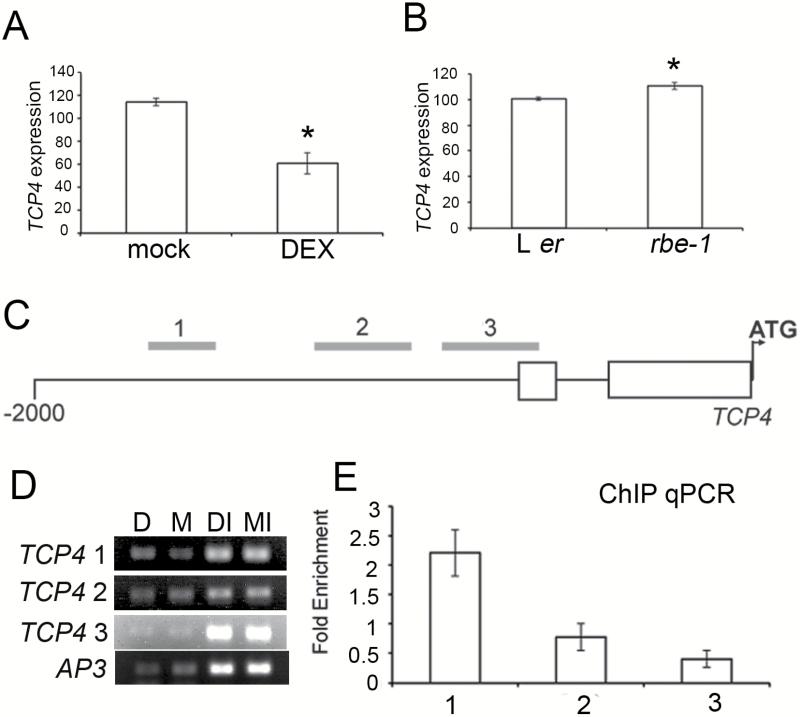
Fig. 1.
RBE negatively regulates TCP4 by directly interacting with its promoter. (A) Expression of TCP4 examined by qRT-PCR in 4 hour DEX- and mock-treated 35S:GR-RBE flowers. Note that TCP4 expression is reduced in DEX-treated flowers. (B) Expression of TCP4 examined by qRT-PCR in L er and rbe-1 flowers. Note that TCP4 expression is increased in rbe-1 compared with L er. In (A) and (B) the y-axis shows relative expression levels normalized to the value of ACT2. Error bars represent stand error of the mean (SEM). Asterisks show a significant difference from the control (P < 0.05, Student t test). (C) Relative positions of fragment 1, 2, and 3 used for ChIP in the -2000 region upstream of the start codon of TCP4. Open boxes indicate 5 prime non-coding exons. (D) Results of semi-quantitative PCR to amplify fragment 1, 2, and 3 in (C) and the control gene (AP3). D: DEX-treated samples; M: mock-treated samples. DI and MI indicate input controls. (E) Fold enrichment of fragment 1, 2 and 3 in (C) in DEX- versus mock-treated flowers assessed by ChIP-qPCR after normalizing with input controls. Fragment 1 is approximately 2.5-fold enriched in DEX- versus mock-treatment, consistent with the semi-qPCR result. Error bars represent SEM for three biological replicates.