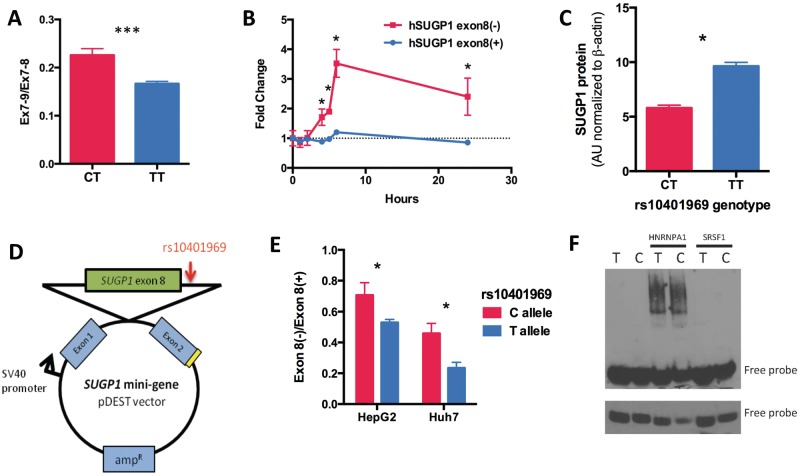
Figure 5.
rs10401969 regulates SUGP1 exon 8 skipping by modulating HNRNPA1 binding. (A) The rs10401969 C allele modulates increases in SUGP1 exon 8 skipping compared to the reference allele by ∼20%. SUGP1 transcript levels were quantified by qPCR in LCLs genotyped for rs10401969 (CT n = 18; TT n = 30). Total SUGP1 was quantified by qPCR assays for exon 7–9 (Ex7-9) and exon 7–8 (Ex7-8) junctions. The relative ratio of Ex7-9 and Ex7-8 is shown. (B) To measure non-sense-medicated mRNA decay, HepG2 cells were treated with cycloheximide (1 μg/ml), and SUGP1 8(−) and 8(+) transcripts were quantified over 24 hours by qPCR. *P < 0.05, two-tailed paired t-test, n = 3. (C) SUGP1 protein levels from CAP LCLs genotyped for the rs10401696 SNP (CT n = 28, TT n = 32) were measured by western blot. Band densities were calculated by Image J and normalized to a β-actin protein levels. (D) Scheme of the SUGP1 mini-gene (mSUGP1) construct containing the genomic fragment of SUGP1 introns 7–8 with the rs10401969 minor allele (C) introduced through site directed mutagenesis. (E) HepG2 and Huh7 (n = 3) cell lines were transfected with either the rs10401969 ‘T’ or ‘C’ allele mSUGP1 construct and mini-gene-derived transcripts quantified by qPCR after 48 hours. Values shown are the relative ratios of mSUGP1 exon 8(−) to mSUGP1 exon 8(+) transcripts are shown. (F) RNA gel shift assay was performed in triplicate with rs10401969 3’ biotin-labeled RNA oligos and either His-tagged HNRNPA1 or SRSF1 proteins. The upper panel shows 30-minute exposure image of a representative gel while the lower panel is a 2-minute exposure of the same gel.