Abstract
Spinal muscular atrophy is a devastating disease that is characterized by degeneration and death of a specific subclass of motor neurons in the anterior horn of the spinal cord. Although the gene responsible, survival motor neuron 1 (SMN1), was identified 20 years ago, it has proven difficult to investigate its effects in vivo. Consequently, a number of key questions regarding the molecular and cellular functions of this molecule have remained unanswered. We developed a Caenorhabditis elegans model of smn-1 loss-of-function using a neuron-specific RNA interference strategy to knock-down smn-1 selectively in a subclass of motor neurons. The transgenic animals presented a cell-autonomous, age-dependent degeneration of motor neurons detected as locomotory defects and the disappearance of presynaptic and cytoplasmic fluorescent markers in targeted neurons. This degeneration led to neuronal death as revealed by positive reactivity to genetic and chemical cell-death markers. We show that genes of the classical apoptosis pathway are involved in the smn-1-mediated neuronal death, and that this phenotype can be rescued by the expression of human SMN1, indicating a functional conservation between the two orthologs. Finally, we determined that Plastin3/plst-1 genetically interacts with smn-1 to prevent degeneration, and that treatment with valproic acid is able to rescue the degenerative phenotype. These results provide novel insights into the cellular and molecular mechanisms that lead to the loss of motor neurons when SMN1 function is reduced.
Introduction
Spinal muscular atrophy (SMA) is an autosomal recessive neurodegenerative disease and one of the most common genetic causes of infant mortality (1). The disease, for which no effective treatment is currently available (2), is characterized by the specific loss of lower spinal cord motor neurons, leading to the atrophy of innervated muscles, paralysis and ultimately death (3). Based on age of onset and motor disabilities, SMA can be clinically subdivided into three types of varying severity (4), all of which are associated with deletions or point mutations in the survival motor neuron 1 gene [SMN1 (5)], which has been mapped to a highly unstable, telomeric region of chromosome 5 (5q13) (6). In addition to SMN1, humans have a nearly identical gene called SMN2; however, the presence of a C-to-T mutation within exon 7 of SMN2 gives rise to a truncated protein, which is rapidly degraded (7). As SMN2 is present in different copy numbers in the genome and is able to produce very low levels of functional protein (5–10% of the total SMN), it acts solely as a genetic modifier when SMN1 is altered, and the level of its expression is responsible for the varying severity of the disease (8).
SMN proteins are ubiquitously expressed both within and outside the nervous system (9). SMN is a component of the well-characterized Gemin complex, which is involved in a series of basic cellular processes, including the biogenesis and assembly of small nuclear ribonucleic proteins and pre-mRNA splicing and transcription (10,11). SMN also plays a role in axonal growth and neuromuscular junction formation, as well as axonal mRNA transport, sub-cellular localization and/or local translation (12,13). However, the specific loss of spinal cord motor neurons in SMA patients cannot be explained simply in terms of their SMN expression pattern, and it is not completely clear which of the SMN functions is responsible for the specific degeneration and death of motor neurons (14).
Modeling SMA in animal systems is therefore crucial to further understand the pathogenesis of the disease and the function played by SMN in motor neurons, as well as to identify potential therapies. Several vertebrate and invertebrate models have been developed, which have provided important information on the changes in the nervous system that occur in response to loss of SMN1 (13,15–17). However, the absence of a second SMN gene in the genome of all animal models, including Caenorhabditis elegans, makes it very difficult to investigate the in vivo role of SMN1 due to the early lethality of SMN1 knock-outs (13,15,16,18–20), hindering the ability to visualize and study this form of neuronal degeneration. In particular, in the current animal models of SMA it has not been possible to determine in vivo the steps that lead to neuron degeneration (13) and, with few exceptions (19,21), it has not been possible to observe neuronal death when SMN1 function is reduced or absent.
Caenorhabditis elegans has a single gene homologous to human SMN, smn-1, which is ubiquitously expressed during all developmental stages (18). The nematode SMN protein exhibits an overall sequence homology and a conserved domain topology with its human counterpart (18). Moreover, the basic functions of SMN, including the ability to self-associate and bind to partner proteins, are conserved in C. elegans (18,22). As observed in other species, a genetic deletion of smn-1 (null allele ok355) causes strong larval lethality (20). The very few surviving ok355 homozygous larvae (escapers) show impaired locomotion and pharyngeal pumping defects, but not neuronal degeneration (20,23). Similar phenotypes have been observed after systemic RNA interference [RNAi (18,22)]. Finally, a hypomorphic mutant allele (cb131) with a single amino acid substitution (D27N) that mimics a point mutation found in some SMA patients (D44V) has been generated (24). Although this mutation overcomes the viability problems of the null mutant, the muscle and nervous system architecture are not affected and there is little change in motility.
Here, we report a novel SMA model in C. elegans. Using cell-specific RNAi knock-down (25), we successfully generated viable transgenic strains in which the function of smn-1 is specifically reduced in a subclass of motor neurons. These transgenic animals mimic some key features of SMA, including impaired locomotion, neuronal degeneration and neuronal death. We report, for the first time, the early steps in neuron degeneration and the loss of neurons due to smn-1 depletion in vivo. We reveal that the neuronal degeneration and death induced by smn-1 reduction are cell-autonomous and proceed in part through an apoptotic mechanism. Furthermore, we investigate the conserved neural protective function of SMN1 from nematodes to humans, and reveal that Plastin3/plst-1 is a genetic interactor of SMN1 in neuronal death. Finally, we show that valproic acid (VPA) is able to rescue the degeneration of C. elegans smn-1-depleted neurons.
Results
Cell-specific knock-down of smn-1 in D-type motor neurons causes movement defects and loss of targeted neurons
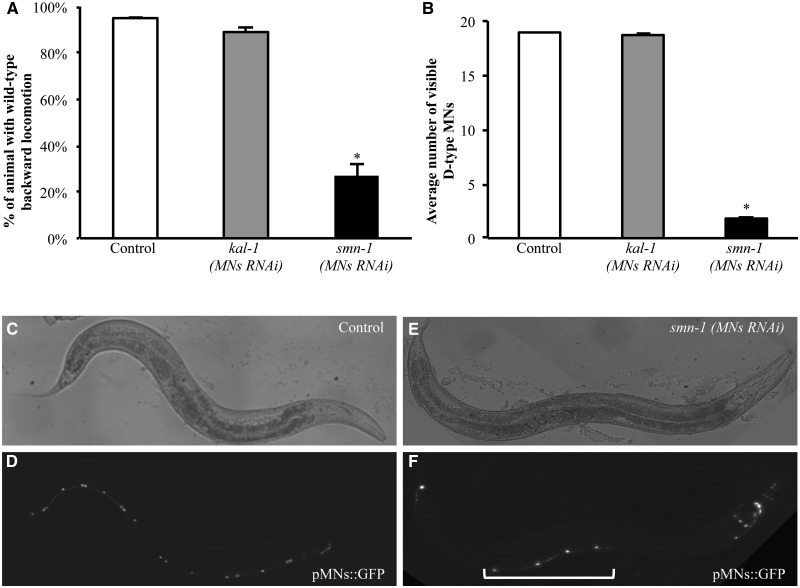
A transgene-driven RNAi strategy (25) was used in C. elegans to knock-down the essential gene smn-1 selectively in the 19 D-type gamma-aminobutyric acid (GABA)ergic motor neurons (punc-25::smn-1 MNs RNAi) (26). The smn-1-silenced strains obtained were viable, fertile, and did not present any visible developmental defects. Given the role of D-type motor neurons in the regulation of C. elegans backward movement (26), we tested this locomotion behavior. As a negative control, we used animals in which we had silenced, under the same conditions and in the same cells, kal-1, a C. elegans neurodevelopmental gene that is not normally expressed in the D-type motor neurons (27). We found that the normal backward response was highly reduced in smn-1 silenced animals (27%), compared with non-silenced controls (95%) and kal-1-silenced animals (89%) (P < 0.001; Fig. 1A and Supplementary Material, Table S1). In contrast, the forward movement was unaffected in all three strains (data not shown), suggesting that the muscles and other classes of motor neurons were not compromised. To confirm that the defect in backward movement was the result of D-type motor neurons dysfunction, we analyzed the thrashing behavior of the smn-1 knocked-down animals that requires functional cholinergic neurons (28). We found that thrashing was not altered in the smn-1 knocked-down animals (Supplementary Material, Fig. S2), indicating that the RNAi had not spread to other tissues.
Figure 1.
smn-1 cell-specific knock-down causes defective backward locomotion and loss of D-type motor neurons cell bodies. (A) Cell-specific knock-down of smn-1 in D-type motor neurons results in alteration of motor neuron functionality. Each bar represents the mean percentage of animals with wild-type backward locomotion ± S.E.M. For each transgene, the results shown are the mean of those obtained from animals of three independent strains; 60 animals were observed for the control strain, and 120 animals were observed for the kal-1(MNs RNAi) and smn-1(MNs RNAi) transgenic strains. * indicates significantly different from control and kal-1(MNs RNAi) (P < 0.005, non-parametric Mann–Whitney test). (B) Average number of motor neurons visible after smn-1(MNs RNAi) knock-down and in negative controls. Each bar represents the mean ± S.E.M. of visible cells and for each transgenic strain the results shown are the mean of those obtained from animals of three independent lines; >150 animals were observed for control strains, and >300 animals were observed for kal-1(MNs RNAi) and smn-1(MNs RNAi) transgenic strains. * indicates significantly different from wild-type and kal-1(MNs RNAi) (P < 0.001, non-parametric Mann–Whitney test). (C–F) oxIs12 [punc-47::GFP] (pMNs::GFP) transgenic animals, with D-type motor neurons expressing GFP in the ventral cord. In the upper panels the animals were observed with visible light, and in the lower panel they were observed with epifluorescence. Anterior is right and ventral is down. In control animals (D) all 19 motor neurons are visible in the ventral cord. smn-1 knock-down (F) induces a reduction in motor neurons, with only four neurons still visible (bracket). The other cells expressing GFP that can be visualized in (F) are other cells expressing the selection marker or neurons other than D-type motor neurons. In these cells, the promoter used to knock-down smn-1 is not expressed. All transgenic strains were obtained using a medium concentration of interfering construct.
We next investigated the effect of smn-1 knock-down on the survival of the 19 D-type motor neurons. We transferred the smn-1 knock-down transgene to a strain that expressed GFP in these cells (oxIs12 [punc-47::GFP]) (Fig. 1D). smn-1 knock-down induced a dramatic reduction in the number of visible GFP-labeled D-type motor neurons, with an average of 1.8 neurons observed per young adult animal (Fig. 1B and F and Supplementary Material, Table S3); no defects were observed in non-silenced controls or in kal-1-silenced animals (average of 19 and 18.7 neurons/animal, respectively; Fig. 1B and D and Supplementary Material, Table S3). A very low but significant reduction of visible motor neurons was also observed after classical systemic RNAi of smn-1 (Supplementary Material, Table S3). We confirmed that the knock-down did not spread to nearby cholinergic motor neurons in the ventral cord by using a strain in which the D-type motor neurons expressed RFP and the cholinergic neurons expressed GFP (vsIs48[punc-17::GFP]; punc-47::RFP). smn-1 knock-down induced a strong reduction of visible D-type motor neurons but no alteration in the number of visible cholinergic neurons (Supplementary Material, Fig. S4 A–C).
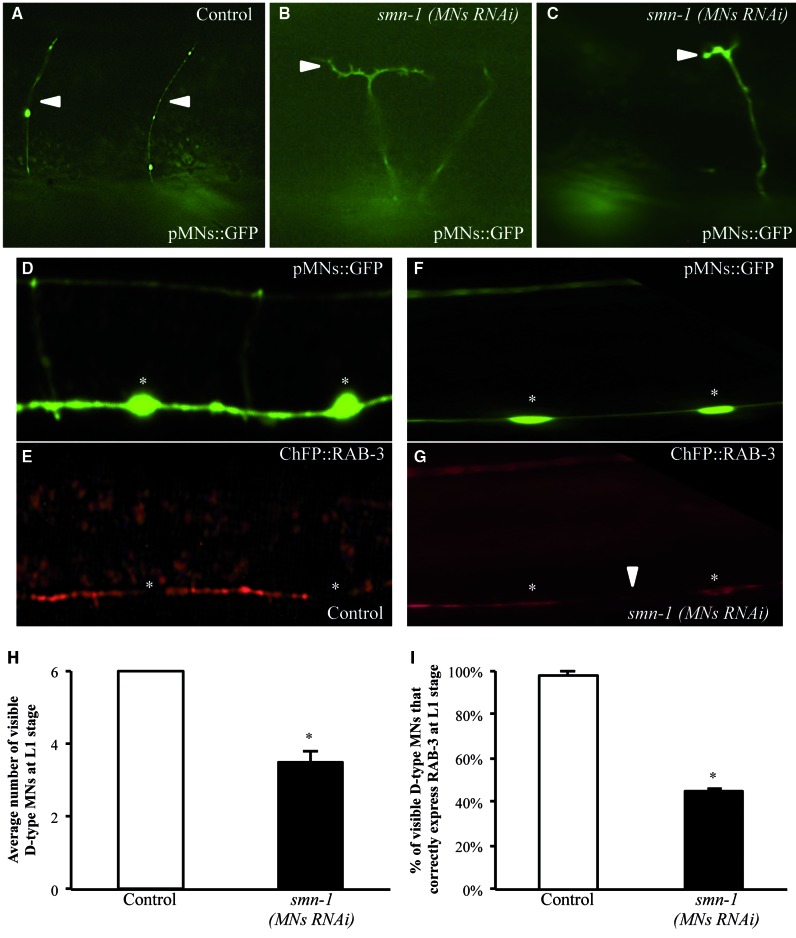
To further characterize this apparent neuronal loss, we investigated the axonal morphology and synaptic distribution of D-type motor neurons, as their alteration could be early signs of a degenerative process. D-type motor neurons are located in the ventral cord and each neuron extends a circumferential commissure to the dorsal side of the animal (26); these axonal processes can be visualized in the transgenic strain described above (oxIs12) (Fig. 2A). In the few surviving GFP-labeled motor neurons of smn-1 silenced animals, we observed a low but significant defect in axon morphology consisting of extra branching and guidance defects (7% defective commissures; P < 0.001) (Fig. 2B and C and Supplementary Material, Table S5). Presynaptic densities can be visualized using the vesicle-associated Ras GTPase, RAB-3, fused to the mCherry fluorescent protein (vdIs4[punc-25::ChFP::RAB-3]) (29). We combined the smn-1(MNs RNAi) with the transgenes to visualize the D-type motor neurons with GFP (oxIs12) and their presynaptic loci with mCherry (vdIs4). In non-silenced animals, RAB-3 was continuously visible in the ventral and the dorsal cord in all animals, with a punctate pattern (Fig. 2E). On the contrary, in 92% of smn-1 knocked-down animals we observed a defect in RAB-3 localization (Fig. 2G and Supplementary Material, Fig. S6). Defective localization of RAB-3 consisted of gaps in the punctate pattern, of different length/severity, and sometimes the complete absence of RAB-3 protein in the entire cord (Supplementary Material, Fig. S6), which suggests a progressive loss of RAB-3 protein at the presynaptic site. To establish whether this defect precedes neuron disappearance, we focused on D-type motor neurons expressing GFP and analyzed RAB-3 localization only in these cells. As very few D-type motor neurons are visible in smn-1 knocked-down animals at the adult stage (1.8 neurons per young adult animal; Fig. 1B and Supplementary Material, Table S3), we analyzed younger animals (L1 stage) that presented an average of 3.5 visible neurons per animal (Fig. 2H). All the GFP-labeled cells expressed RAB-3 in L1 non-silenced animals (Fig. 2I). In contrast, in smn-1 knocked-down animals only 45% of the remaining GFP-labeled neurons still correctly expressed the RAB-3 marker. These results reveal that the earliest observable defect of smn-1 knock-down is a reduced number of presynaptic densities, followed by morphological alterations in the axon and loss of cytoplasmic expression of GFP.
Figure 2.
smn-1 RNAi knock-down causes defects in D-type motor neurons axons and synapse. (A–C) RNAi knock-down of smn-1 in D-type motor neurons produces changes in motor neuron axonal morphology. In oxIs12 (pMNs::GFP) transgenic animals motor neuron axons are visible. Anterior is right and ventral is down. In control animals (A) commissures are directed to the dorsal site (up) as single axons (arrowheads). smn-1 knock-down (B, C) induces axonal branching and defects in axonal guidance. Transgenic strains were obtained using a medium concentration of interfering construct. (D–G) vdIs4 (pMNs::ChFP::RAB-3), oxIs12 (pMNs::GFP) double transgenic animals allow visualization of the localization of RAB-3, a presynaptic marker, in the ventral cord (E,G) and the motor neuron cytoplasm (D,F), respectively. White asterisks indicate motor neuron cell bodies, where RAB-3 is not localized. Anterior is left and ventral is down. In the upper panels the ventral cord is observed with epifluorescence using a GFP filter, and in the lower panel it is observed using a Texas Red filter. (E) Localization of RAB-3 in motor neurons in control animals. (G) Localization of RAB-3 in animals knocked-down for smn-1. A small gap of presynaptic RAB-3 (white arrowhead) is visible in the ventral cord, where axons are still present in the corresponding area in the upper panel. (H) Average number of motor neurons visible after smn-1(MNs RNAi) knock-down and in controls at L1, the first larval stage after hatching. At this stage, six D-type neurons express GFP in the wild-type animals. Each bar represents the mean number of visible cells ± S.E.M.; >30 animals were observed. * indicates significantly different from control (P < 0.005, non-parametric Mann–Whitney test). (I) Percentage of visible motor neurons that correctly express RAB-3 after smn-1(MNs RNAi) and in the control strain ± S.E.M. >30 animals were observed. * indicates significantly different from control (P < 0.001, non-parametric z statistic test). Transgenic strains in (F–I) were obtained using a high concentration of interfering construct.
smn-1 knock-down causes neuronal death through an apoptotic mechanism
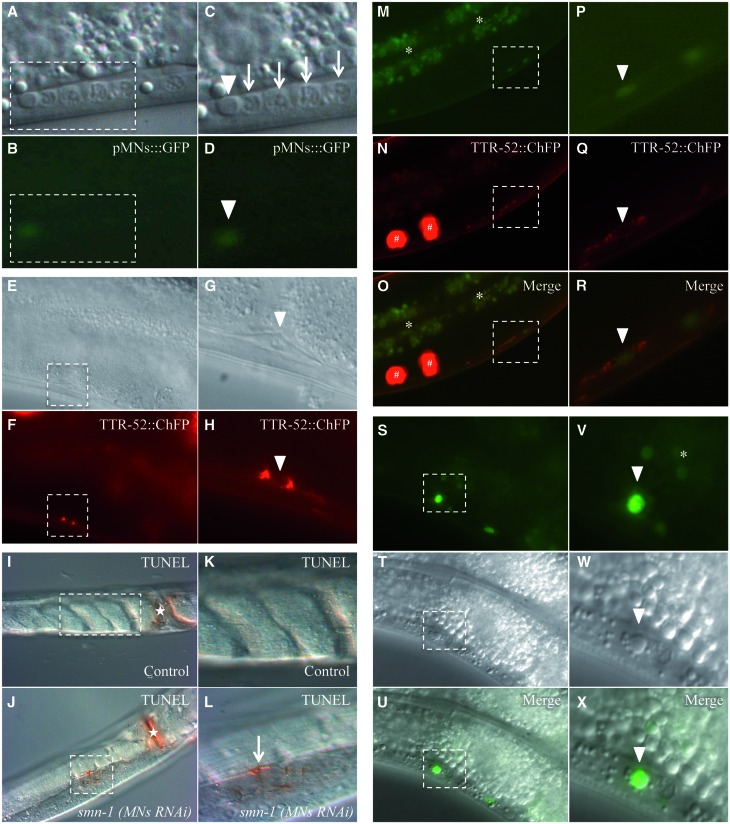
To determine whether loss of GFP corresponded to cell death, we first investigated whether apoptosis could be detected using Differential Interference Contrast (DIC) microscopy. In C. elegans, programmed cell death causes a characteristic change in cell morphology and refractility that can be observed in living animals using DIC microscopy (30,31). This unique feature made it possible to genetically dissect for the first time the programmed cell death pathway in C. elegans (30). We found that in smn-1(MNs RNAi) animals, some neurons lacking GFP expression presented an altered morphology of the nucleus, with a button-like shape characteristic of apoptotic corpses (Fig. 3A and C and Supplementary Material, Table S7A). Occasionally, the apoptotic appearance was also observed in neurons that were still faintly GFP fluorescent (Fig. 3B and D). A further marker of cell death is the presence on the surface of dying cells of ‘eat-me’ signals recognized by engulfing cells. One such signal is the transthyretin-like secreted protein TTR-52 (32). We took advantage of a transgenic strain expressing TTR-52 fused to the red fluorophore mCherry (smIs119[phsp-16.2::TTR-52::ChFP]), which specifically labels apoptotic dying cells before engulfment (32). In smn-1(MNs RNAi); smIs119 double transgenic animals, we observed TTR-52-positive signals with the typical ring shape in the nerve cord (Fig. 3F and H and Supplementary Material, Table S7B). The fragmentation of chromosomal DNA is a late event of apoptosis and can be visualized using the TUNEL assay. This assay revealed few positive spots in the ventral nerve cord when smn-1 was knocked-down (Fig. 3J and L).
Figure 3.
The degeneration of motor neurons after smn-1 knock-down ends with the death of the cell and accumulation of fluorescence. The motor neuron death induced by smn-1(MNs RNAi) can be visualized in four different ways. (A–D) Using DIC microscopy in the ventral cord of smn-1 transgenic animals we observed a button-like structure typical of apoptotic dying cells (white arrowheads) in association with a neuron faintly expressing GFP from the oxIs12 (pMNs::GFP) transgene. Arrows correspond to viable neuronal nuclei in the ventral cord. (C, D) Enlargements of areas outlined in (A) and (B). Upper images were obtained with visible light, and lower images with GFP filter epifluorescence. (E–H) A TTR-52::ChFP positive neuron, with a typical ring shape, is observed in the ventral cord of smn-1(MNs RNAi), smIs119 (TTR-52::ChFP) double transgenic animals (white arrowheads). A quantification of TTR-52::ChFP positive neurons is reported in Supplementary Material, Table S7. (G, H) Enlargements of areas outlined in (E) and (F). Upper panels were obtained with visible light, and lower panels were obtained with Texas Red filter epifluorescence. (I–L) Apoptotic dying neurons in the ventral cord were analyzed in permeabilized animals using TUNEL assay. smn-1(MNs RNAi) knocked-down animals, but not controls, presented TUNEL-reactive cells along the ventral cord. (I) Control animal, adult stage, presenting no labeling in the cord. (K) Enlargement of the area outlined in I. (J) smn-1(MNs RNAi) animal, adult stage, presenting a positively labeled cell in the cord. (L) Enlargement of the area outlined in J, showing the labeled cell in the cord (arrow). n > 40 for control and for smn-1(MNs RNAi). White asterisks correspond to the vulva, which is always labeled. All images were taken with visible light and DIC. (M–R) Accumulation of fluorescence is sometimes surrounded by a TTR-52::ChFP ring (white arrowheads), in smn-1(MNs RNAi), smIs119 (TTR-52::ChFP) double transgenic animals. (P–R) Enlargements of areas outlined in (M)–(O). In the upper panels images were taken with epifluorescence using a GFP filter; in the middle panels a Texas Red filter was used, and the lower panels are merge of the two images. (S–X) Accumulation of fluorescence sometimes corresponds to a button-like structure, using DIC microscopy, typical of apoptotic dying cells (white arrowheads). A quantification of neurons accumulating fluorescence is reported in Figure 4A and in Supplementary Material, Table S7. (V–X) Enlargements of areas outlined in (S)–(U). In the upper panels images were taken with epifluorescence using a GFP filter; the middle panels are DIC images, and in the lower panels are merge of the two images. White asterisks correspond to intestinal autofluorescence and white hashtags to coelomocytes, scavenger cells that phagocyte secreted TTR-52::ChFP. Anterior is right and ventral is down in all images, except in (I)–(L) which are ventral views. All transgenic strains were obtained using an HC of interfering construct.
These findings, which were never observed in control animals (Fig. 3I and K and Supplementary Material, Table S7B), indicate that smn-1 knock-down causes the degeneration and death of D-type motor neurons, at least in part through an apoptotic mechanism.
smn-1-silenced dying D-type motor neurons acquire fluorescence
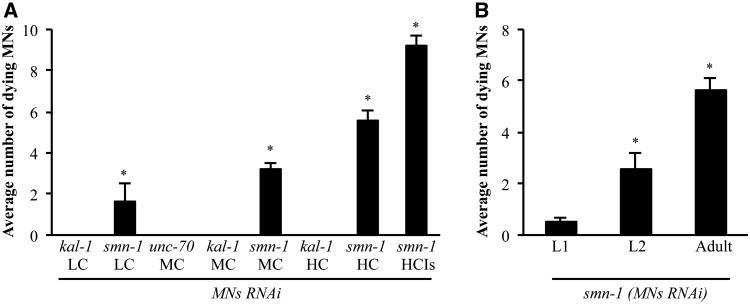
While analyzing smn-1 silenced animals carrying the smIs119[phsp-16.2::TTR-52::ChFP] transgene, we unexpectedly observed a faint but clear and reproducible accumulation of green fluorescence in D-type motor neurons (Fig. 3M and P). These animals did not carry any reporter expressing GFP in neurons. In some cases, the red fluorescent ‘eat-me’ signal of the TTR-52::ChFP transgene surrounded a cell that was accumulating green fluorescence (Fig. 3M–R). Moreover, some of these cells presented the refractile, button-like structure typical of apoptotic dying cells (Fig. 3S–X). The smn-1 knock-down transgenic strain carried as the selection marker a GFP reporter (GB301 pchs-2::gfp), which is specifically expressed in 17 cell nuclei in the pharynx (33). This reporter is never expressed in D-type motor neurons. Analysis of the emission spectra of the dying fluorescent neurons in the nerve cord with confocal microscopy and immunostaining against GFP unambiguously revealed that the fluorescence observed in these cells was due to GFP (data not shown). A similar acquired green fluorescence was observed when we used a different selection marker expressing GFP in the intestinal nuclei (pJM67 pelt-2::gfp) (34) (Supplementary Material, Table S8). Finally, acquired red fluorescence was observed when we used the pchs-2 promoter fused to the red fluorophore DsRed2 (GB300 pchs-2::DsRed2) (Supplementary Material, Table S8). This novel-acquired fluorescence could in principle be due to an uptake by motor neurons of the fluorophore produced by different cells or, more likely, to a gene deregulation in the dying neurons that promotes the leakage expression of the fluorophore from the promoter used as the marker. Although the precise mechanism underlying this phenotype has not yet been elucidated, the accumulation of fluorescent material in D-type motor neurons correlated with their degeneration induced by the knock-down of smn-1. We therefore decided to determine whether this fluorescence could be used as a read-out for smn-1-induced neuronal death, as it would greatly facilitate any further investigation with this strain. To confirm the specificity of this phenotype, we silenced smn-1 in a dose-dependent manner by using different concentrations of interfering transgene (25). At concentrations of 200 ng/μl (high), 10 ng/μl (medium) and 2 ng/μl (low), we observed an average of 5.2, 3.2 and 1.0 green fluorescent neurons per animal in the ventral nerve cord, respectively (high concentration (HC), medium concentration (MC) and low concentration (LC) in Fig. 4A and Supplementary Material, Table S8). Finally, by integrating the knock-down transgene in the genome, the acquired fluorescence was maintained and had the highest penetrance (9.2 green fluorescent neurons per animal), as expected from loss of mosaicism following integration (HC integrated (HCIs) in Fig. 4A and Supplementary Material, Table S8) (35). Importantly, this phenotype was never observed in control animals in which kal-1 had been silenced at similar concentrations (Fig. 4A and Supplementary Material, Table S8). As an additional negative control, we silenced a gene known to play a function in D-type motor neurons (unc-70), but whose knock-down causes various abnormalities but not cell death (36); in this case, we also never observed fluorescent neurons (Fig. 4A and Supplementary Material, Table S8). To establish the timing of the acquired fluorescence, we correlated this phenotype to the presynaptic defect. We focused on D-type dying motor neurons and analyzed RAB-3 localization in these neurons in smn-1 knocked-down animals. Among 460 neurons presenting acquired fluorescence in knocked-down animals (n = 99 animals), 386 (84%) presented a complete absence of RAB-3 protein, strongly suggesting that in almost all cases the fluorescence is acquired after RAB-3 is lost at the presynaptic sites (data not shown).
Figure 4.
Quantification and temporal characterization of motor neuron death induced by smn-1(MNs RNAi). (A) Accumulation of fluorescence in D-type motor neurons in the ventral cord of smn-1(MNs RNAi) animals is dependent on the transgene dose. Number of motor neurons accumulating fluorescence in transgenic animals. Bars represent the average number of fluorescence accumulating neurons per animal and for each transgenic strain the results shown are the mean of those obtained from animals of at least two independent line ± S.E.M.; >280 animals were observed for kal-1(MNs RNAi LC), >500 animals were observed for smn-1(MNs RNAi LC), 100 animals were observed for each transgenic strain at MC, >150 animals were observed for each transgenic strain at HC, and 100 animals were observed for the smn-1(MNs RNAi) integrated line (HCIs). * indicates significantly different from unc-70(MNs RNAi) and kal-1(MNs RNAi) (P < 0.001, non-parametric Mann–Whitney test). (B) The number of motor neurons accumulating fluorescence increases with ageing. Bars represent the average number of fluorescence-accumulating neurons per animal. L1 is the first larval stage after hatch. L2 is the subsequent larval stage. >20 animals were observed at the L1 stage, 50 animals were observed at the L2 stage, and 130 animals were observed at the adult stage. * indicates significantly different from previous larval stage (P < 0.001, non-parametric Mann–-Whitney test). Transgenic strains were obtained using an HC of interfering construct.
Degeneration and death of D-type motor neurons are age-dependent and can be rescued by human SMN1
SMA is a genetic disease showing a progressive neuron degeneration following an early onset of the disease. Therefore, we investigated whether after smn-1 knock-down the observed phenotypes worsened with age. We found that loss of motor neurons could already be observed during the first larval stage (L1) post-hatching (Fig. 2H), and significantly increased in the following larval stage (L2) (data not shown) and in adults (Fig. 1B and Supplementary Material, Table S3). The accumulation of acquired fluorescence in the ventral cord neurons began to be visible after hatching (L1), and progressively increased with age (Fig. 4B). Thus, loss of motor neurons after smn-1 knock-down is a progressive phenomenon, a finding consistent with the progression of the disease.
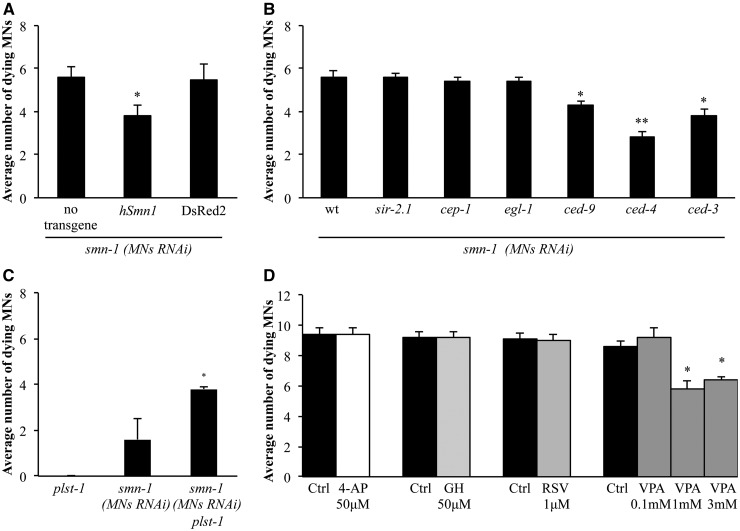
The high level of conservation in the domain topology and sequence of SMN in C. elegans and humans (36% similarity) (18) prompted us to determine whether their function was also conserved. To address this point, we performed a rescue experiment in which the wild-type copy of the human SMN1 cDNA was expressed pan-neuronally (punc-119::hSMN1) in SMN-1 knocked-down animals. We observed a significant reduction in the number of dying neurons in these animals compared with wild-type animals (3.8 versus 5.6 fluorescent neurons per animal, P < 0.001) (Fig. 5A and Supplementary Material, Table S8), indicating a rescue of neuronal death using the human copy of the gene. We also found that the SMN1 human gene could rescue, at least partially, the phenotypes of reduced life span, thrashing, and backward movement, observed in the smn-1 null mutant ok355 (Supplementary Material, Fig. S9A–D).
Figure 5.
Genetic and pharmacological characterization of motor neuron death induced by smn-1(MNs RNAi). (A) Expression of the full length form of human SMN1 cDNA using a pan-neuronal promoter partially rescues the cell death phenotype induced by smn-1(MNs RNAi). Bars represent the average number of fluorescence-accumulating neurons per animal and for each transgenic strain the results shown are the mean of those obtained from animals of at least two independent lines. In total, 130 animals were observed for the control (no transgene overexpression), and >380 animals were observed for both the human (hSMN1) and DsRed2 overexpressing strains. * indicates significantly different from smn-1(MNs RNAi) (P < 0.001, non-parametric Mann–Whitney test) and from overexpression of DsRed2 in similar conditions. In all graphs the S.E.M. is shown. Transgenic strains were obtained using an HC of interfering construct. (B) Apoptotic genes were tested for their involvement in smn-1(MNs RNAi) induced motor neuron degeneration. A significant reduction of the number of fluorescence-accumulating motor neurons was observed using a gain-of-function mutation in ced-9 and loss-of-function mutations in ced-4 and ced-3. Bars represent the average number of GFP-accumulating neurons per animal. In total, 130 animals were observed for smn-1(MNs RNAi), and >200 animals were observed for smn-1(MNs RNAi) in the sir-2.1(ok434), cep-1(gk138), egl-1(n1084n3082), or ced-9(n1950) background; 100 animals were observed for smn-1(MNs RNAi) in the ced-4(n1162) or ced-3(n717) background. * indicates significantly different from smn-1(MNs RNAi) in the wild-type background (P < 0.001, non-parametric Mann–Whitney test); ** indicates significantly different from smn-1(MNs RNAi) in the wild-type background (P < 0.001, non-parametric Mann–Whitney test) and significantly different from smn-1(MNs RNAi) in the ced-9 (P < 0.001, non-parametric Mann–Whitney test) or ced-3 mutant backgrounds (P < 0.02, non-parametric Mann–Whitney test). Transgenic strains were obtained using an HC of interfering construct. (C) A loss of function mutation in plst-1(tm4255) enhances the motor neuron death induced by smn-1(MNs RNAi). All three strains express the fluorescent marker pchs-2::gfp. Bars represent the average number of fluorescence-accumulating neurons per animal. >250 animals were observed for plst-1(tm4255), >500 animals were observed for smn-1(MNs RNAi), and >150 animals were observed for smn-1(MNs RNAi) in the plst-1(tm4255) background. * indicates significantly different from smn-1(MNs RNAi) in the wild-type background (P < 0.001, non-parametric Mann–Whitney test). Transgenic strains were obtained using an LC of interfering construct. (D) VPA was able to partially rescue smn-1(MNs RNAi) induced degeneration; it reduced the number of fluorescence-accumulating neurons at 1 and 3 mm. 4-AP, GH, resveratrol (RSV) had no effect. Bars represent the average number of fluorescence-accumulating neurons per animal. >100 animals were observed for each treatment. * indicates significantly different from smn-1(MNs RNAi) exposed to buffer alone (P < 0.001, unpaired t-test). In all graphs the S.E.M. is shown. Transgenic strains were obtained using an HC of interfering integrated construct.
Taken together, these data support a significant functional conservation between the human and C. elegans genes, and confirm that the motor neuron death observed when smn-1 is knocked-down is the result of a specific reduction in the SMN-1 protein.
smn-1 knock-down leads to the activation of the cell-death pathway
Having implied an apoptotic process in our findings, we next tested animals carrying mutations in genes of the canonical apoptotic pathway, including egl-1, ced-9, ced-4 and ced-3 (37–39), as well as cep-1, which encodes an ortholog of the human tumor suppressor p53 (40), and sir-2.1, a member of the Sirtuin family (41). The latter two genes act upstream of the apoptotic core machinery through two parallel mechanisms activated after DNA damage, and p53 has been demonstrated to interact with SMN1 in cell cultures (42). We transferred the smn-1 interfering construct at high concentration (HC) into six individual strains, each mutated in one of these genes. With the exception of ced-9, all the mutations were loss-of-function to determine whether lack of their pro-apoptotic role prevented cell death. ced-9 is normally protective, and a gain-of-function mutation was used to determine a possible role in preventing smn-1-induced cell death. Using the acquired fluorescence in dying motor neurons as a readout, we observed a significant reduction in the number of fluorescent neurons in the smn-1 knocked-down strain (5.6 per animal) when in combination with a gain-of-function mutation in ced-9(n1950) (4.3 per animal, P < 0.001) or the loss-of-function mutations in ced-4(n1162) (2.8 per animal, P < 0.001), or ced-3(n717) (3.8 per animal, P < 0.001) (Fig. 5B). In contrast, loss-of-function mutations in cep-1(gk138), sir-2.1(ok434), and surprisingly also egl-1(n1084 n3082), did not significantly affect the number of dying motor neurons after smn-1 knock-down. When the smn-1 knock-down transgene was not in the background, we never observed fluorescent dying motor neurons in ced-9(n1950) gain-of-function mutants or ced-4(n1162) and ced-3(n717) loss-of-function mutants (Supplementary Material, Table S8). These results confirm that motor neurons undergo apoptotic cell death involving some of the key components of the classical pathway, and indicate that the accumulation of fluorescence in smn-1-silenced motor neurons is a late event in the degeneration process that partially occurs after the activation of the cell death pathway genes.
smn-1-induced degeneration is enhanced by mutations in Plastin3 and rescued by VPA
One of the main advantages of the cell-specific smn-1 knock-down animal models is that the use of different concentrations of the interfering construct makes it possible to mimic an allelic series with phenotypes of different severity. This facilitates the identification of genetic elements and pharmacological molecules that can either enhance or ameliorate the observed phenotypes. Plastin3 is an actin-bundling protein that has been reported to exert specific protective effects in SMA patients, as higher expression levels correlate with milder clinical symptoms (43). In addition, work in different species suggests that Plastin3 is a cross-species modifier of SMN function (43,44). Given that the C. elegans plst-1 gene encodes a protein similar to Plastin3 (56% identity and 73% similarity), we tested whether lack of plst-1 acts as a genetic enhancer of degeneration induced by smn-1 knock-down. When we introduced the plst-1(tm4255) loss-of-function mutation in the LC transgenic model, we observed that it significantly increased the number of dying motor neurons (from 1.6 to 3.8 dying neurons per animal, P < 0.001) (Fig. 5C). This result shows that Plastin3 plays a role in preventing neuronal death in vivo, and that smn-1 genetically interacts with Plastin3 in this process.
We next asked whether selected candidate drugs could rescue the degeneration phenotype. Resveratrol, VPA, 4-aminopyridine (4-AP) and gaboxadol (GH) have been shown to have a protective effect in different models of neuron degeneration (4,24,45). We found that, in the integrated strain of smn-1 knock-down, VPA significantly reduced the number of dying neurons (Fig. 5D). The extent of rescue was similar at 1 and 3 mM, but absent at 0.1 mM. These results demonstrate that VPA has a protective effect on neurons affected by smn-1 mutation.
Discussion
Although the genetic basis of SMA and the molecular functions of the SMN protein have been extensively studied, several fundamental questions remain largely unanswered (46). How does the absence of SMN1, a ubiquitous gene, induce the selective degeneration of motor neurons? Is the degeneration of motor neurons in SMA cell-autonomous or secondary to muscle defects? Do primarily developmental defects play a significant role in triggering the degeneration of neurons? The novel C. elegans model of SMA that we have presented here overcomes many of the limitations of other available genetic models (20,24), allowing us to study in vivo the selective effects of the loss of SMN1 function in motor neurons. Our transgenic animals, harboring knock-down of smn-1 selectively in D-type motor neurons, are healthy and easy to maintain, eliminating the pleiotropic phenotypes that often complicate the interpretation of motor neuron alterations in genetic models based on the systemic loss of smn-1 function. Our results support the notion that the loss of D-type motor neuron function is caused by degeneration and death of these cells, with no spreading to nearby cholinergic neurons in the nerve cord; this is in agreement with the idea that degeneration of motor neurons is a cell-autonomous phenomenon resulting from the cell-specific knock-down of smn-1. Finally, our data on different developmental stages of the animal provide additional evidence that loss of motor neurons is an age-dependent process, with degeneration and neuronal death progressively increasing as the animals get older. This result suggests that the knock-down of smn-1 is triggering a degenerative process, whereas developmental defects appear to play a minor role in the neuronal phenotypes we observe.
Degeneration culminates in neuronal death partially through an apoptotic mechanism
Motor neuron degeneration is the primary hallmark of SMA. The earliest morphological sign of degeneration that we observe is the loss of presynaptic densities: the gaps in the RAB-3 punctate pattern in the nerve cord are already present before the loss of visible GFP, and before the appearance of more severe signs of degeneration such as refractility of the cell body and positivity to cell death markers. Remarkably, neuronal death was not reported in a previous study in C. elegans of the null mutant smn-1(ok355) (20), and in our experiments with systemic smn-1 RNAi we observed neuronal loss at much lower penetrance than in smn-1(MNs RNAi) animals. These results can be explained by the hypothesis that maternal contribution largely prevents neuronal degeneration in the very few adult animals that survive past the early larval stages. This may be one of the reasons why neuron degeneration and death were not reported in previous studies using Drosophila melanogaster, zebrafish and C. elegans models of SMA (13,20,24,47).
SMN1-induced neuronal death could in principle occur through a necrotic or an apoptotic process. Our experiments conclusively demonstrate that the degeneration process in the smn-1(MNs RNAi) animals can activate an apoptotis-like cell death, whereas we never observed necrotic cells, which are easily distinguishable using DIC microscopy (data not shown). We have shown that the nuclei of dying neurons assume in some cases a button-like shape and become refractile and TUNEL-positive. Their cell bodies also bind TTR-52, a signal for engulfment that is pivotal for the clearance of dead neurons. Furthermore, we have identified part of the molecular pathway activated by smn-1 knock-down, revealing that it includes ced-9, ced-4 and ced-3 of the core apoptotic machinery, but not cep-1, sir-2.1 and egl-1. This result excludes the involvement of a DNA damage-induced apoptotic pathway (41). Our results reveal that neuronal death is independent of EGL-1 and that CED-9 has a minor role, confirming the findings of previous studies in which induction of apoptosis can occur downstream from, or in parallel with, egl-1 and ced-9 (48) (Supplementary Material, Fig. S10). Moreover, CED-9-dependent cell death pathways that are EGL-1-independent have already been reported in C. elegans (49). However, we never observed the complete suppression of the cell-death phenotype, even when apoptotic genes were absent. The presence of residual cell death even in ced-9, ced-4 and ced-3 mutants suggests that the apoptotic pathway is only one of the cell death mechanisms activated after smn-1 knock-down, and that it may require additional caspase or non-apoptotic/alternative death mechanisms (50,51). We therefore suggest that CED-4/Apaf-1 is also activated by an alternative upstream mechanism and that CED-4/Apaf-1 itself activates other downstream effectors, in addition to CED-3/ICE-caspase (Supplementary Material, Fig. S10). Indeed, CED-4-dependent cell death pathways that are CED-3-independent have already been reported in yeast strains, in which CED-4 is overexpressed (52), and in C. elegans (49–51), for example, after RNAi knock-down of the presumptive anti-apoptotic protein, ICD-1 (53). Moreover CoQ depletion triggers a cell death mechanism in D-type motor neurons that is independent of EGL-1 and CED-9, but requires CED-4 and CED-3, with CED-4 also activating other downstream effectors in addition to CED-3 (54). Thus, the downstream genes alternative to CED-3 remains to be elucidated, although these may include other members of the caspase family or novel components.
Accumulation of GFP as a new read-out for dying neurons
Neuronal degeneration is detectable as the accumulation of GFP in dying motor neurons, an unexpected and novel phenotype whose specific dependence on the knock-down of smn-1 has been confirmed using cell-death markers and genetic mutants. GFP accumulation was a late event in neuronal degeneration, as (i) it occurred after loss of RAB-3 and after the activation of the cell death pathway genes, (ii) it was associated with indicators of cell death, and (iii) the number of GFP-positive neurons was very low in the early larval stages and strongly increased with age. We still do not know the mechanism by which GFP accumulates in dying motor neurons. It is possible that transcription from chs-2 or other promoters in dying motor neurons, where chs-2 is normally not active, is not tightly controlled such that small amounts of gfp transcripts are made and translated. An alternative mechanism is that GFP, expressed in the appropriate cells under the chs-2 promoter, is partially released in the pseudocoelom and taken up by degenerating/dying neurons in the ventral cord, where it accumulates. Independent of the origin, the accumulation of GFP is a specific, late sign of the degeneration of the motor neurons in which smn-1 has been knocked-down, and is never observed in negative controls. Further studies are needed to elucidate the mechanism of accumulation and whether or not this phenomenon can also be used as a marker of other neuronal apoptotic deaths.
Plastin3 interaction with SMN1 and the axonal theory of SMA pathogenesis
Plastin3, an actin-bundling protein, was the first documented modifier of SMA and a high level of its expression has been shown to have a protective effect in patients (43). Plastin3 has also been shown to be a cross-species modifier of SMN function. Overexpression of Plastin3 rescues the defects associated with SMN1 down-regulation in motor neurons of SMA mouse embryos and in a zebrafish model of SMA (43,55). Mutation of Fimbrin, the Drosophila ortholog of Plastin3, exacerbates DmSmn loss-of-function defects (44). When the C. elegans ortholog, plst-1, is knocked-down it causes unclear and opposite effects on growth defects and on pharyngeal pumping alterations observed in smn-1(ok355) mutants (44). We have now demonstrated that the loss of function of plst-1 enhances the degeneration and neuronal death of motor neurons in smn-1(MNs RNAi) animals, confirming a conserved neuronal protective role of this protein. The protective role of plst-1, together with the early loss of presynaptic densities in smn-1(MNs RNAi) animals, suggests that SMN1 loss-of-function in axons and at synaptic terminals could underscore the pathophysiology of SMA. It has been suggested that the specific association between SMN1 and a subset of mRNAs and proteins in the axonal compartment could be pivotal for the maintenance of axons and synapses in motor neurons. According to this hypothesis, known as the ‘axonal theory’ of SMA pathogenesis (56), this process could be selectively impaired in SMA, leading to the degeneration and loss of neurons in affected patients. The alterations at the presynaptic terminals may trigger a ‘die back’ mechanism that eventually results in neuronal death. These results suggest that the specific role of smn-1 in the presynaptic subcellular compartment requires a deeper analysis.
Uses of the SMA model for pharmacological and genetic screenings
Pharmacological screenings of SMA drugs, using cellular models and a few available animal models of SMA, including C. elegans (24), have already been performed with interesting results. However, in most of the existing animal models degeneration and death of motor neurons either are not present or cannot be easily monitored in vivo (19,20). The novel, viable model of neuron degeneration that we describe is particularly suited for approaches aimed at identifying molecules specifically capable of protecting against neuronal degeneration and death. The small group of chemicals tested here was chosen for their ability to improve the deficits in locomotion and fitness caused by loss of function of smn-1 in C. elegans, or their proven protective role in other C. elegans models (4,24,45). We obtained a significant rescue with the anticonvulsant VPA, a deacetylase inhibitor. VPA has been associated with an increase in SMN genes expression in cell cultures and has been successfully used in mouse SMA models (4,46). Our results reveal that VPA can antagonize the neuronal death caused by smn-1 depletion in vivo, confirming the relevance of our model for drug screenings. The results of clinical trials investigating the potential of VPA as an SMA therapeutic have been inconsistent, demonstrating that it is only beneficial in a restricted subset of patients. Our results lend support to the use of VPA in clinical trials and our model could also play an important role in deciphering factors that influence the individual response to this drug. Interestingly, the two molecules, 4-AP and GH, that rescued the fitness defects in previous C. elegans smn-1 mutants (24), were not effective in our model, confirming that the cell-specific RNAi approach developed here can reveal new molecules that are specifically important for neuron degeneration and not for the general functions of smn-1.
There are two additional points worth mentioning regarding the value of the cell-specific knock-down of smn-1. First, this approach offers the opportunity to analyze the effects of smn-1 depletion in different neuronal sub-classes and/or in other non-neuronal tissues, with the goal of ultimately understanding the basis of the specificity of motor neuron loss observed in SMA patients. Second, by using different concentrations of the smn-1 interfering construct it is possible to modulate the extent of the cell-specific knock-down, and we have indeed obtained different lines presenting phenotypes of increasing severity, similar to a genetic allelic series. This has allowed us to test genetic mutations and drugs expected to suppress (e.g. ced-3, VPA) or worsen (e.g. plst-1) the phenotypes of smn-1(MNs RNAi) animals (Fig. 5B–D). Complementary models presenting different penetrance of the defects could be used to establish screens to identify genetic modifiers of smn-1 knock-down capable of either enhancing or suppressing neuronal degeneration and death. This could help to elucidate the molecular mechanisms underlying SMA and therefore its pathogenesis, and drive the discovery of new drug targets for this disease.
Materials and Methods
Strains
Nematodes were grown and handled following standard procedures, under uncrowded conditions, at 20 °C, on NGM (nematode growth medium) agar plates seeded with Escherichia coli strain OP50 (57). Wild-type animals were C. elegans variety Bristol, strain N2. The alleles and transgenic strains used in this work were: unc-55(e1170) I, rrf-3(pk1426) II, smn-1(ok355) I, ced-4(n1162) III, sir-2.1(ok434) IV, ced-3(n717) IV, cep-1(gk138) I, ced-1(e1735) I, ced-2(e1752) IV, oxIs12[punc-47::GFP; lin-15(+)] X, juIs76[punc-25::GFP; lin-15(+)] II and vsIs48[punc-17::GFP] provided by the Caenorhabditis Genetics Center (CGC), which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440); Is[punc-47::RFP] kindly provided by K. Shen (Stanford University, USA); plst-1(tm4255) provided by S. Mitani, through the National Bio-Resource Project of the MEXT, Japan; smIs119[phsp-16.2::TTR-52::ChFP; KSII(+); unc-76(+)] kindly provided by D. Xue (University of Colorado, USA); egl-1(n1084n3082) and ced-9(n1950) kindly provided by M. Hengartner (University of Zurich, Switzerland). The transgenes obtained in this work were: gbEx29[punc-25::smn-1 RNAi HC], gbEx571[punc-25::smn-1 RNAi MC], gbEx502[punc-25::smn-1 RNAi LC], gbEx566[punc-25::kal-1 RNAi LC], gbEx625[punc-25::kal-1 RNAi MC], gbEx543[punc-25::kal-1 RNAi HC], gbEx588[punc-25::unc-70 RNAi MC], gbEx529[punc-119::hSMN1], gbEx540[punc-119::DsRed2], gbEx580[punc-25::smn-1 RNAi HC], gbEx581[punc-25::smn-1 RNAi HC], gbEx612[pchs-2::GFP], gbIs4[punc-25::smn-1 RNAi HC], vdIs4[punc-25::ChFP::RAB-3]. A full description of the transgenes is available in the Supplementary Material, Supplementary Methods section. Genetic crosses were made to transfer transgenes to the appropriate genetic background. In all cases, the presence of the mutant allele was verified by polymerase chain reaction (PCR) followed, when necessary, by sequencing. Primer sequences are available on request. Two independent clones with the same genotype were examined after each cross (see Supplementary Material, Supplementary Tables) and the mean of the two clones has been reported in the results.
Neuron-specific RNAi knock-down
D-type motor neuron-specific RNAi transgenes were constructed as previously described (25) by PCR-fusing a D-type motor neuron-specific promoter to an exon-rich region of the target gene (58). To knock-down a gene of interest we used a short form of unc-25/GAD promoter (180 bp), which is specifically expressed from embryonic to adult stages in the 19 GABAergic D-type motor neurons in the ventral cord and not in other GABAergic neurons (RME, AVL, DVB and RIS) (59). We confirmed the expected expression pattern of this promoter using a gfp reporter approach, and successfully tested the ability of this promoter to knock-down a control gene, specifically in D-type motor neurons (data not shown). For the genes to be knocked down we amplified, from genomic DNA, the same exon-rich region that was used for the RNAi plasmid library, prepared for RNAi by feeding experiments (60). Exon-rich regions were amplified in two separate PCR reactions to obtain the sense and antisense fragments. The unc-25 promoter was amplified using specific primers. The promoter was subsequently fused to each orientation of the target gene by PCR fusion using internal primers, as previously described (25,58). Primer sequences are available on request.
Transgenic lines
GB300 [pchs-2::NLS::DsRed2] plasmid was obtained by removing the chs-2 promoter from GB301 [pchs-2::NLS::GFP] (33) and inserting it in front of the coding sequence of pVH14.05 (promoterless DsRed2 vector, kindly provided by H. Hutter, Simon Fraser University, Burnaby, Canada) using SalI and BamHI restriction sites. pSM punc-25::ChFP::RAB-3 was generated by removing the unc-25 promoter from pNV punc-25::GFP and inserting it in front of the coding sequence in the plasmid pNV ChFP::RAB-3, using FseI and AscI restriction sites. Germline transformation was performed as described previously (61). Sense and antisense fusions to the GABAergic promoter were pooled in equimolar amounts and microinjected at different concentrations. HC, MC and LC of knocking-down constructs were injected, which corresponded to 200, 10 and 2 ng/μl, respectively. The LC knock-down construct was injected in rrf-3 (pk1426) mutants, which are more sensitive to neuronal RNAi, to enhance the knock-down effects (61). pSM punc-25::ChFP::RAB-3 plasmid was injected together with an empty pSM plasmid to obtain vdEx410 [pSM punc-25::ChFP::RAB-3; pSM; pSM podr-1::GFP]. vdEx410 and gbEx29 transgenic lines were integrated by TMP/UV treatment to obtain vdIs4 and gbIs4, respectively. The co-injection markers used in this work were: podr-1::GFP (kindly provided by C. Bargmann, The Rockfeller University, New York, USA; GFP expression in AWC and AWB neurons), pJM371 pelt-2::NLS::RFP and pJM67 pelt-2::NLS::GFP (kindly provided by J. McGhee, University of Calgary, Canada; RFP and GFP expression in the intestinal nuclei), GB301 pchs-2::NLS::GFP and GB300 pchs-2::NLS::DsRed2 (GFP and DsRed2 expression in 17 nuclei of the pharynx). The injection markers were co-injected at 10 to 30 ng/μl (see Supplementary Material, Supplementary Methods). At least two transgenic lines were examined for each experiment (see Supplementary Material, Supplementary Methods) and the mean of the lines has been reported in the figures. All transgenic strains are listed in the Supplementary Material, Supplementary Methods.
Behavioral assay
Well-fed, young adult animals were used for backward movement assay to test D-type motor neurons function. The assay was performed blind on NGM plates, 6 cm in diameter, seeded with bacteria. Using an eyelash the animal was touched first on the tail to induce a forward movement and then on the head to test for backward movement. Defective movement was scored when animals were unable to fully move backward. For each data set, the percentage of defective animals among the total number of tested animals was calculated.
TUNEL assay
Young adult and adult animals were fixed and permeabilized as previously described for GABA neurotransmitter visualization (62). Terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) reactions were then performed by using an In Situ Cell Death Detection Kit, POD (Roche Diagnostics). Briefly, permeabilized animals were washed twice with PBS and incubated with 50 μl of TUNEL reaction mixture for 60 min at 37 °C. For visualization of samples under a light microscope, the animals were washed three times with PBS before adding 50 μl of Converter-POD. The animals were incubated for 30 min at 37°C and then rinsed three times with PBS. Finally, for signal conversion, 100 μl of DAB substrate (Sigma-Aldrich) was added to each sample, followed by incubation at room temperature for 10 min. Animals were washed three times with PBS before mounting on slides and analyzing under a light microscope.
Microscopy analysis
Animals were immobilized in 0.01% tetramisole hydrochloride (Sigma-Aldrich) on 4% agar pads and visualized using Zeiss Axioskop or Leica DMI6000B microscopes. All microscopes were equipped with epifluorescence and DIC Nomarski optics and images were collected with an Axiocam digital camera or with Leica digital cameras DFC 480 and 420 RGB. To discriminate dying motor neuron fluorescence from endogenous autofluorescence, we used either a Zeiss filter set 09 (that allowed observation of intestinal cell autofluorescence in yellow and GFP-positive dying cells in green) on a Zeiss Axioskop microscope or a Leica TCS SP2 AOBS laser scanning confocal microscope.
Rescue experiments
The rescue construct for pan-neuronal expression of hSMN1 was created by PCR-fusion (58) of three fragments: the promoter of unc-119 gene, the cDNA of the human SMN1 gene and the 3′ untranslated region (UTR) of unc-54. The unc-119 promoter has been previously used for neuronal-directed rescue of smn-1 (20) and is considered to be a strong transcriptional inducer (63). The human SMN1 gene was amplified from plasmid pcDNA4::hSMN1. To increase the stability of the rescuing construct, we added the 3′ UTR of the unc-54 gene (which was amplified from plasmid pPD95.75, kindly provided by A. Fire, Stanford University, Stanford, USA) to the 3′ end of the human cDNA. This construct was microinjected at two concentrations (20 and 100 ng/μl) to find the best conditions and first injected in wild-type animals to exclude non-specific effects due to overexpression of the gene (data not shown). The same transgenic array was then transferred to animals in which smn-1 was knocked-down in the motor neurons. As a negative control the human SMN1 gene was replaced with DsRed2 (amplified from plasmid pVH14.05), a gene not present in the C. elegans genome. The specificity of the rescue with the human gene was confirmed by the lack of rescue when DsRed2 was expressed in smn-1 knocked-down animals, using the same experimental conditions (Fig. 5A and Supplementary Material, Table S8).
Drug treatments
Individual compounds (Sigma Aldrich) were dissolved in dimethylsulfoxide (Sigma Aldrich) or H2O and then added to molten NGM agar to the desired concentration before pouring into petri dishes. Control NGM plates contained the appropriate dilution of vehicle. Plates were allowed to dry for 1 day before seeding with OP50 bacteria, after which they were incubated at room temperature. Gravid adults were then transferred onto the drug/control plates to lay eggs and removed after 24 h. Each experimental condition was run in blind duplicate or triplicate. For drug treatments, the integrated line with an HC of interfering construct (HCIs) was used so that all animals tested were affected due to the loss of incomplete segregation of the transgene that is obtained following integration.
Statistical analysis
Primer of Biostatistics and GraphPad Prism softwares were used for statistical analyses. The statistical significance was determined using z-statistic tests, unpaired t-test or Mann–Whitney test, as indicated, comparing each sample against the control. The standard error of the mean was used to estimate variation within a single population.
Supplementary Material
Supplementary Material is available at HMG online.
Acknowledgements
The authors thank Rowan Tweedale for critical reading of the manuscript and useful suggestions; M. Hengartner (University of Zurich, Switzerland), D. Xue (University of Colorado, USA), J. McGhee (University of Calgary, Canada), K. Shen (Stanford University, USA), C.I. Bargmann (The Rockfeller University, New York, USA), H. Hutter (Simon Fraser University, Burnaby, Canada) and A. Fire (Stanford University, USA) for reagents; the IGB Open Laboratory for facilities and resources; the IGB Integrated Microscopy and the PhD program in Molecular and Cellular Biotechnology of the Second University of Naples for support; the Caenorhabditis Genetics Center (CGC), which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440), the C. elegans Gene Knockout Project at OMRF and the C. elegans Reverse Genetics Core Facility at U.B.C., which are part of the International C. elegans Gene Knockout Consortium, and the MITANI Laboratory, through the National Bio-Resource Project of the MEXT, Japan, for mutant strains; Wormbase.
Conflict of Interest statement. None declared.
Funding
Italian Ministry of Economy and Finance (grant Project FaReBio di Qualità); Italian Ministry of Health (RF2009-1473235). This work was partially supported by the Italian Ministry of Instruction University and Research (FIRB-Merit grant number RBNE08LN4P_002) to E.D.S.; the Institute of Genetics and Biophysics PhD fellowship program to I.G.; the Compagnia di San Paolo (2008.1224) to S.C.; NHMRC (Project Grants 1067461, and SRF Fellowship APP1111042) and ARC (Future Fellowship FT110100097) to M.A.H.; University of Queensland, Postgraduate Award to J.C.; University of Queensland, International Postgraduate Award to A.D. Funding to pay the Open Access publication charges for this article was provided by Italian Ministry of Economy and Finance and Italian Ministry of Health.
References
- 1.Pearn J. (1978) Incidence, prevalence, and gene frequency studies of chronic childhood spinal muscular atrophy. J. Med. Genet., 15, 409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seo J., Howell M.D., Singh N.N., Singh R.N. (2013) Spinal muscular atrophy: an update on therapeutic progress. Biochim. Biophys. Acta, 1832, 2180–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crawford T.O., Pardo C.A. (1996) The neurobiology of childhood spinal muscular atrophy. Neurobiol. Dis., 3, 97–110. [DOI] [PubMed] [Google Scholar]
- 4.Pearn J. (1980) Classification of spinal muscular atrophies. Lancet, 1, 919–922. [DOI] [PubMed] [Google Scholar]
- 5.Lefebvre S., Burglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M. et al. (1995) Identification and characterization of a spinal muscular atrophy-determining gene. Cell, 80, 155–165. [DOI] [PubMed] [Google Scholar]
- 6.Brzustowicz L.M., Lehner T., Castilla L.H., Penchaszadeh G.K., Wilhelmsen K.C., Daniels R., Davies K.E., Leppert M., Ziter F., Wood D. et al. (1990) Genetic mapping of chronic childhood-onset spinal muscular atrophy to chromosome 5q11.2-13.3. Nature, 344, 540–541. [DOI] [PubMed] [Google Scholar]
- 7.Monani U.R., Lorson C.L., Parsons D.W., Prior T.W., Androphy E.J., Burghes A.H., McPherson J.D. (1999) A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum. Mol. Genet., 8, 1177–1183. [DOI] [PubMed] [Google Scholar]
- 8.Vitali T., Sossi V., Tiziano F., Zappata S., Giuli A., Paravatou-Petsotas M., Neri G., Brahe C. (1999) Detection of the survival motor neuron (SMN) genes by FISH: further evidence for a role for SMN2 in the modulation of disease severity in SMA patients. Hum. Mol. Genet., 8, 2525–2532. [DOI] [PubMed] [Google Scholar]
- 9.Young P.J., Le T.T., thi Man N., Burghes A.H., Morris G.E. (2000) The relationship between SMN, the spinal muscular atrophy protein, and nuclear coiled bodies in differentiated tissues and cultured cells. Exp. Cell. Res., 256, 365–374. [DOI] [PubMed] [Google Scholar]
- 10.Pellizzoni L., Yong J., Dreyfuss G. (2002) Essential role for the SMN complex in the specificity of snRNP assembly. Science, 298, 1775–1779. [DOI] [PubMed] [Google Scholar]
- 11.Fischer U., Liu Q., Dreyfuss G. (1997) The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell, 90, 1023–1029. [DOI] [PubMed] [Google Scholar]
- 12.Rossoll W., Jablonka S., Andreassi C., Kroning A.K., Karle K., Monani U.R., Sendtner M. (2003) Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of beta-actin mRNA in growth cones of motoneurons. J. Cell. Biol., 163, 801–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan Y.B., Miguel-Aliaga I., Franks C., Thomas N., Trulzsch B., Sattelle D.B., Davies K.E., van den Heuvel M. (2003) Neuromuscular defects in a Drosophila survival motor neuron gene mutant. Hum. Mol. Genet., 12, 1367–1376. [DOI] [PubMed] [Google Scholar]
- 14.Burghes A.H., Beattie C.E. (2009) Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat. Neurosci., 10, 597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang H.C., Dimlich D.N., Yokokura T., Mukherjee A., Kankel M.W., Sen A., Sridhar V., Fulga T.A., Hart A.C., Van Vactor D. et al. (2008) Modeling spinal muscular atrophy in Drosophila. PLoS One, 3, e3209.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boon K.L., Xiao S., McWhorter M.L., Donn T., Wolf-Saxon E., Bohnsack M.T., Moens C.B., Beattie C.E. (2009) Zebrafish survival motor neuron mutants exhibit presynaptic neuromuscular junction defects. Hum. Mol. Genet., 18, 3615–3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imlach W.L., Beck E.S., Choi B.J., Lotti F., Pellizzoni L., McCabe B.D. (2012) SMN is required for sensory-motor circuit function in Drosophila. Cell, 151, 427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miguel-Aliaga I., Culetto E., Walker D.S., Baylis H.A., Sattelle D.B., Davies K.E. (1999) The Caenorhabditis elegans orthologue of the human gene responsible for spinal muscular atrophy is a maternal product critical for germline maturation and embryonic viability. Hum. Mol. Genet., 8, 2133–2143. [DOI] [PubMed] [Google Scholar]
- 19.Schrank B., Gotz R., Gunnersen J.M., Ure J.M., Toyka K.V., Smith A.G., Sendtner M. (1997) Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc. Natl Acad. Sci. U.S.A., 94, 9920–9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Briese M., Esmaeili B., Fraboulet S., Burt E.C., Christodoulou S., Towers P.R., Davies K.E., Sattelle D.B. (2009) Deletion of smn-1, the Caenorhabditis elegans ortholog of the spinal muscular atrophy gene, results in locomotor dysfunction and reduced lifespan. Hum. Mol. Genet., 18, 97–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monani U.R., Sendtner M., Coovert D.D., Parsons D.W., Andreassi C., Le T.T., Jablonka S., Schrank B., Rossoll W., Prior T.W. et al. (2000) The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn(−/−) mice and results in a mouse with spinal muscular atrophy. Hum. Mol. Genet, 9, 333–339. [DOI] [PubMed] [Google Scholar]
- 22.Burt E.C., Towers P.R., Sattelle D.B. (2006) Caenorhabditis elegans in the study of SMN-interacting proteins: a role for SMI-1, an orthologue of human Gemin2 and the identification of novel components of the SMN complex. Invertebr. Neurosci., 6, 145–159. [DOI] [PubMed] [Google Scholar]
- 23.Dimitriadi M., Kye M.J., Kalloo G., Yersak J.M., Sahin M., Hart A.C. (2013) The neuroprotective drug riluzole acts via small conductance Ca2+-activated K+ channels to ameliorate defects in spinal muscular atrophy models. J. Neurosci., 33, 6557–6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sleigh J.N., Buckingham S.D., Esmaeili B., Viswanathan M., Cuppen E., Westlund B.M., Sattelle D.B. (2011) A novel Caenorhabditis elegans allele, smn-1(cb131), mimicking a mild form of spinal muscular atrophy, provides a convenient drug screening platform highlighting new and pre-approved compounds. Hum. Mol. Genet., 20, 245–260. [DOI] [PubMed] [Google Scholar]
- 25.Esposito G., Di Schiavi E., Bergamasco C., Bazzicalupo P. (2007) Efficient and cell specific knock-down of gene function in targeted C. elegans neurons. Gene, 395, 170–176. [DOI] [PubMed] [Google Scholar]
- 26.McIntire S.L., Jorgensen E., Kaplan J., Horvitz H.R. (1993) The GABAergic nervous system of Caenorhabditis elegans. Nature, 364, 337–341. [DOI] [PubMed] [Google Scholar]
- 27.Rugarli E.I., Di Schiavi E., Hilliard M.A., Arbucci S., Ghezzi C., Facciolli A., Coppola G., Ballabio A., Bazzicalupo P. (2002) The Kallmann syndrome gene homolog in C. elegans is involved in epidermal morphogenesis and neurite branching. Development, 129, 1283–1294. [DOI] [PubMed] [Google Scholar]
- 28.Pierce-Shimomura J.T., Chen B.L., Mun J.J., Ho R., Sarkis R., McIntire S.L. (2008) Genetic analysis of crawling and swimming locomotory patterns in C. elegans. Proc. Natl Acad. Sci. U.S.A., 105, 20982–20987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nonet M.L. (1999) Visualization of synaptic specializations in live C. elegans with synaptic vesicle protein-GFP fusions. J. Neurosci. Methods, 89, 33–40. [DOI] [PubMed] [Google Scholar]
- 30.Sulston J.E., Horvitz H.R. (1977) Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol., 56, 110–156. [DOI] [PubMed] [Google Scholar]
- 31.Sulston J.E., Schierenberg E., White J.G., Thomson J.N. (1983) The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol., 100, 64–119. [DOI] [PubMed] [Google Scholar]
- 32.Wang X., Li W., Zhao D., Liu B., Shi Y., Chen B., Yang H., Guo P., Geng X., Shang Z. et al. (2010) Caenorhabditis elegans transthyretin-like protein TTR-52 mediates recognition of apoptotic cells by the CED-1 phagocyte receptor. Nat. Cell. Biol., 12, 655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Veronico P., Gray L.J., Jones J.T., Bazzicalupo P., Arbucci S., Cortese M.R., Di Vito M., De Giorgi C. (2001) Nematode chitin synthases: gene structure, expression and function in Caenorhabditis elegans and the plant parasitic nematode Meloidogyne artiellia. MGG, 266, 28–34. [DOI] [PubMed] [Google Scholar]
- 34.Fukushige T., Hendzel M.J., Bazett-Jones D.P., McGhee J.D. (1999) Direct visualization of the elt-2 gut-specific GATA factor binding to a target promoter inside the living Caenorhabditis elegans embryo. Proc. Natl Acad. Sci. U.S.A., 96, 11883–11888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stinchcomb D.T., Shaw J.E., Carr S.H., Hirsh D. (1985) Extrachromosomal DNA transformation of Caenorhabditis elegans. Mol. Cell. Biol., 5, 3484–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hammarlund M., Jorgensen E.M., Bastiani M.J. (2007) Axons break in animals lacking beta-spectrin. J. Cell. Biol., 176, 269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Conradt B., Horvitz H.R. (1998) The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell, 93, 519–529. [DOI] [PubMed] [Google Scholar]
- 38.Yan N., Gu L., Kokel D., Chai J., Li W., Han A., Chen L., Xue D., Shi Y. (2004) Structural, biochemical, and functional analyses of CED-9 recognition by the proapoptotic proteins EGL-1 and CED-4. Mol. Cell, 15, 999–1006. [DOI] [PubMed] [Google Scholar]
- 39.Yang X., Chang H.Y., Baltimore D. (1998) Essential role of CED-4 oligomerization in CED-3 activation and apoptosis. Science, 281, 1355–1357. [DOI] [PubMed] [Google Scholar]
- 40.Derry W.B., Putzke A.P., Rothman J.H. (2001) Caenorhabditis elegans p53: role in apoptosis, meiosis, and stress resistance. Science, 294, 591–595. [DOI] [PubMed] [Google Scholar]
- 41.Greiss S., Hall J., Ahmed S., Gartner A. (2008) C. elegans SIR-2.1 translocation is linked to a proapoptotic pathway parallel to cep-1/p53 during DNA damage-induced apoptosis. Gene Dev., 22, 2831–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Young P.J., Day P.M., Zhou J., Androphy E.J., Morris G.E., Lorson C.L. (2002) A direct interaction between the survival motor neuron protein and p53 and its relationship to spinal muscular atrophy. J. Biol. Chem., 277, 2852–2859. [DOI] [PubMed] [Google Scholar]
- 43.Oprea G.E., Krober S., McWhorter M.L., Rossoll W., Muller S., Krawczak M., Bassell G.J., Beattie C.E., Wirth B. (2008) Plastin 3 is a protective modifier of autosomal recessive spinal muscular atrophy. Science, 320, 524–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dimitriadi M., Sleigh J.N., Walker A., Chang H.C., Sen A., Kalloo G., Harris J., Barsby T., Walsh M.B., Satterlee J.S. et al. (2010) Conserved genes act as modifiers of invertebrate SMN loss of function defects. PLoS Genet., 6, e1001172.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parker J.A., Arango M., Abderrahmane S., Lambert E., Tourette C., Catoire H., Neri C. (2005) Resveratrol rescues mutant polyglutamine cytotoxicity in nematode and mammalian neurons. Nat. Genet., 37, 349–350. [DOI] [PubMed] [Google Scholar]
- 46.Sumner C.J. (2007) Molecular mechanisms of spinal muscular atrophy. J. Child Neurol., 22, 979–989. [DOI] [PubMed] [Google Scholar]
- 47.McWhorter M.L., Monani U.R., Burghes A.H., Beattie C.E. (2003) Knockdown of the survival motor neuron (Smn) protein in zebrafish causes defects in motor axon outgrowth and pathfinding. J. Cell Biol., 162, 919–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwartz H.T., Horvitz H.R. (2007) The C. elegans protein CEH-30 protects male-specific neurons from apoptosis independently of the Bcl-2 homolog CED-9. Gene Dev., 21, 3181–3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yee C., Yang W., Hekimi S. (2014) The intrinsic apoptosis pathway mediates the pro-longevity response to mitochondrial ROS in C. elegans. Cell, 157, 897–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blum E.S., Driscoll M., Shaham S. (2008) Noncanonical cell death programs in the nematode Caenorhabditis elegans. Cell Death Differ., 15, 1124–1131. [DOI] [PubMed] [Google Scholar]
- 51.Shaham S. (1998) Identification of multiple Caenorhabditis elegans caspases and their potential roles in proteolytic cascades. J. Biol. Chem., 273, 35109–35117. [DOI] [PubMed] [Google Scholar]
- 52.Tao W., Walke D.W., Morgan J.I. (1999) Oligomerized Ced-4 kills budding yeast through a caspase-independent mechanism. Biochem. Biophys. Res. Commun., 260, 799–805. [DOI] [PubMed] [Google Scholar]
- 53.Bloss T.A., Witze E.S., Rothman J.H. (2003) Suppression of CED-3-independent apoptosis by mitochondrial betaNAC in Caenorhabditis elegans. Nature, 424, 1066–1071. [DOI] [PubMed] [Google Scholar]
- 54.Earls L.R., Hacker M.L., Watson J.D., Miller D.M., III (2010) Coenzyme Q protects Caenorhabditis elegans GABA neurons from calcium-dependent degeneration. Proc. Natl Acad. Sci. U.S.A., 107, 14460–14465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ackermann B., Krober S., Torres-Benito L., Borgmann A., Peters M., Hosseini Barkooie S.M., Tejero R., Jakubik M., Schreml J., Milbradt J. et al. (2013) Plastin 3 ameliorates spinal muscular atrophy via delayed axon pruning and improves neuromuscular junction functionality. Hum. Mol. Genet., 22, 1328–1347. [DOI] [PubMed] [Google Scholar]
- 56.Carrel T.L., McWhorter M.L., Workman E., Zhang H., Wolstencroft E.C., Lorson C., Bassell G.J., Burghes A.H., Beattie C.E. (2006) Survival motor neuron function in motor axons is independent of functions required for small nuclear ribonucleoprotein biogenesis. J. Neurosci., 26, 11014–11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brenner S. (1974) The genetics of Caenorhabditis elegans. Genetics, 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hobert O. (2002) PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. BioTechniques, 32, 728–730. [DOI] [PubMed] [Google Scholar]
- 59.Eastman C., Horvitz H.R., Jin Y. (1999) Coordinated transcriptional regulation of the unc-25 glutamic acid decarboxylase and the unc-47 GABA vesicular transporter by the Caenorhabditis elegans UNC-30 homeodomain protein. J. Neurosci., 19, 6225–6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kamath R.S., Fraser A.G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M. et al. (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature, 421, 231–237. [DOI] [PubMed] [Google Scholar]
- 61.Mello C.C., Kramer J.M., Stinchcomb D., Ambros V. (1991) Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J., 10, 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Duerr J.S. (2006) Immunohistochemistry. WormBook, ed. The C. elegans Research Community, doi/10.1895/wormbook.1.105.1, http://www.wormbook.org. [Google Scholar]
- 63.Maduro M., Pilgrim D. (1995) Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics, 141, 977–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.