Abstract
Proliferating cell nuclear antigen (PCNA) forms a trimeric ring that encircles duplex DNA and acts as an anchor for a number of proteins involved in DNA metabolic processes. PCNA has two structurally similar domains (I and II) linked by a long loop (inter-domain connector loop, IDCL) on the outside of each monomer of the trimeric structure that makes up the DNA clamp. All proteins that bind to PCNA do so via a PCNA-interacting peptide (PIP) motif that binds near the IDCL. A small protein, called TIP, binds to PCNA and inhibits PCNA-dependent activities although it does not contain a canonical PIP motif. The X-ray crystal structure of TIP bound to PCNA reveals that TIP binds to the canonical PIP interaction site, but also extends beyond it through a helix that relocates the IDCL. TIP alters the relationship between domains I and II within the PCNA monomer such that the trimeric ring structure is broken, while the individual domains largely retain their native structure. Small angle X-ray scattering (SAXS) confirms the disruption of the PCNA trimer upon addition of the TIP protein in solution and together with the X-ray crystal data, provides a structural basis for the mechanism of PCNA inhibition by TIP.
INTRODUCTION
Proliferating cell nuclear antigen (PCNA) is an essential protein in archaea and eukarya (1–4). PCNA plays essential roles in nucleic acid metabolic processes including replication, repair, recombination and cell cycle progression (5,6). PCNA is trimeric and forms a ring that encircles duplex DNA. In the trimer, the polypeptides are positioned in a head-to-tail arrangement with the interface between subunits forming an antiparallel β-sheet that is also supported by ionic and hydrophobic interactions [reviewed in: (2)]. Each monomer is composed of two structurally similar domains linked by a long loop (inter-domain connector loop, IDCL) on the outside of each monomer of the trimeric structure (7). When encircling DNA, PCNA interacts with a large number of enzymes and regulates their biochemical properties (6). PCNA primarily interacts with proteins via a PCNA-interacting peptide (PIP) motif (8). The canonical PIP motif is a weak consensus sequence of eight amino acids QXXhXXaa where ‘h’ is a moderately hydrophobic amino acid and ‘a’ is an aromatic residue (9). The PIP-containing proteins bind to the PCNA trimer in a small hydrophobic cavity near the IDCL [reviewed in: (10)].
Genes encoding for PCNA have been identified in all archaeal genomes [(11) and references therein]. While the genomes of most euryarchaeota contain a single gene encoding PCNA, the genome of Thermococcus kodakarensis contains two genes encoding PCNA homologs (TK0535 and TK0582, encoding PCNA1 and PCNA2, respectively) (12–14). Although the two proteins are similar in structure and biochemical properties (12–14), only PCNA1 is essential for T. kodakarensis viability (13,14). We will refer to PCNA1 as PCNA in the remainder of this manuscript.
Using a genetic approach, a small protein that interacts with T. kodakarensis PCNA was previously identified (15). The protein, encoded by TK0808, was shown to inhibit PCNA-dependent activities (16). The gene encoding for the protein is found only in Thermococcales genomes and therefore it was designated TIP (Thermococcales inhibitor of PCNA). TIP is a 64 amino acid protein with a molecular mass of 7.6 kDa and a pI of 9.9 (Figure 1A). The three-dimensional (3D) structure of TIP bound to PCNA was determined using X-ray crystallography. TIP alone is partially disordered (Supplementary Figure S1), but upon binding to PCNA becomes structured. Several residues of TIP form a short helix that binds in a similar manner to other PIP-containing proteins, while the residues that follow form a helix that binds to a hydrophobic region on PCNA that is created by displacement of the IDCL. This movement of the IDCL also alters the relationship of the two domains of the PCNA monomer such that the usual trimeric ring structure cannot form. Small angle X-ray scattering (SAXS) confirms the disruption of the PCNA trimer in the PCNA:TIP complex. The implications of these results for a mechanism of PCNA inhibition in archaea are discussed.
Figure 1.
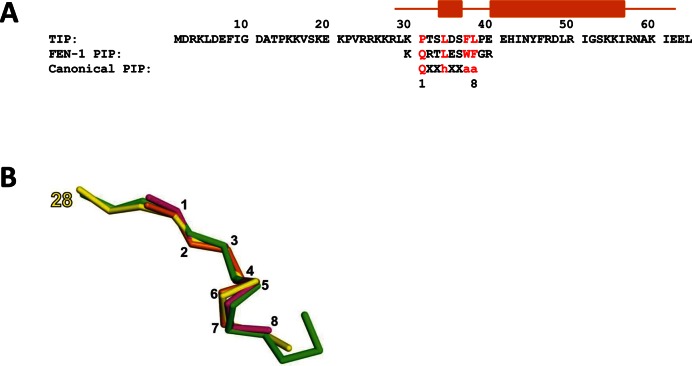
TIP protein contains a non-canonical PIP motif. (A) Amino acid sequence of the TIP protein and alignment to FEN-1 PIP and the consensus PIP motif. Key amino acids in the PIP sequences are in red. Orange bars above the sequence show the position of the helices of TIP in the PCNA:TIP structure. (B) The structural comparison of PIP motif peptides is displayed as the Cα trace of TIP residues 28–39 (PDB ID: 5DA7) in yellow superimposed with PIP site peptides from PCNA:FEN-1 (PDB ID: 5DAI) in pink, DNA polymerase from structure PDB ID: 3A2F (45) in green and RFC from structure PDB ID: 1ISQ (42) in gold. The yellow label marks the starting amino acid in TIP, while the black numbers correspond to residues in the canonical PIP motif. The rms between TIP and the other PIP peptides is ∼0.8 Å.
MATERIALS AND METHODS
Protein and peptides
PCNA protein was expressed and purified as previously described (12). Full length TIP and TIP peptide (residues 28–64) and flap endonuclease 1 (FEN-1) peptide (residues 330–340) were chemically synthesized as follows. The peptides were synthesized on a Tribute peptide synthesizer (Protein Technologies, Inc.) at the 50 μmol scale. Coupling was performed for all amino acids for 20 min, using 1:1:2 0.05 M amino acid:0.05 M HCTU (2-(6-Chloro-1H-benzotriazole-1-yl)-1,1,3,3-tetramethylaminium hexafluorophosphate):0.4 M N-methylmorpholine in DMF (dimethylformamide), with a 10-fold excess of amino acid and activator relative to the Fmoc-Leu-Wang Resin (Novabiochem). Deprotection was performed with 20% piperidine in DMF. Protecting groups for Fmoc synthesis included OtBu (Asp, Glu, Ser, Thr and Tyr), Trt (His and Asn), Boc (Lys) and Pbf (Arg). All amino acids were purchased from Novabiochem.
Following synthesis, the peptides were cleaved from the resin with 88:5:5:2 TFA:water:EDT:TIPS (trifluoroacetic acid:water:1,2-ethanedithiol:triisopropylsilane) for two hours at room temperature. Cold ether was then added to the vial and the peptides were precipitated by centrifugation (10 min, 5500 g); the cold ether precipitation was repeated two times. The samples were dried with nitrogen overnight. The crude peptide was then dissolved in 5% acetonitrile, 0.1% TFA in water and purified on a Shimadzu HPLC equipped with an XSelect CSH C18 Prep column (5 μm, 10 × 250 mm). Samples were run on a gradient from 0 to 100% B and 100 to 0% A, where Buffer A is 0.1% TFA, 5% acetonitrile in water and Buffer B was 95% acetonitrile, 0.1% TFA in water for 60 min at a flow rate of 1 ml/min. UV detection was set for 214 and 280 nm. Mass analysis of the peak eluting at 53–56% Buffer B was performed on a Bruker microflex MALDI-TOF. Appropriate fractions were combined and lyophilized on a Labconco Freezone benchtop lyophilizer.
Crystallization
Crystals of PCNA with TIP were grown by hanging drop vapor diffusion. The ratio of PCNA to TIP was ∼1:1.2. The stock solution of PCNA was 3.2 mg/ml (0.11 mM as monomer) in 500 mM NaCl, 20 mM Tris–HCl (pH 7.0), 0.5 mM ethylenediaminetetraacetic acid and 10% glycerol. The stock solution of TIP was 8 mg/ml (1.05 mM) in 50 mM Tris–HCl (pH 8.0) and 500 mM NaCl. The well solution was 1.2 M ammonium sulfate, 50 mM sodium citrate buffer (pH 4.0) and 10% glycerol. The drops were formed by adding equal volumes of protein and well solution.
For the PCNA:FEN-1 structure, a peptide consisting of 11-residues of the T. kodakarensis FEN-1 protein [residues 330–340 (Figure 1A), encoded by the TK1281 gene] was used. Crystals of PCNA with FEN-1 peptide were grown by sitting drop vapor diffusion. The ratio of PCNA to peptide was approximately 1:5. The PCNA protein was in the same solution as above. The peptide concentration was 2 mM in water. The well solution was 2.8 M ammonium sulfate, 100 mM citric acid buffer (pH 5.3) and 10% polyethylene glycol 4000.
X-ray crystal data collection
The diffraction data for PCNA:FEN-1 peptide, PCNA:TIP peptide and initial data for PCNA:TIP were collected using a Rigaku Micro Max 007 rotating anode generator and a Rigaku RAXIS IV++ detector (Rigaku/MSC, The Woodlands, TX, USA). The crystals were cooled to 100 K with a cryocooler (Cryo Industries, Manchester, NH, USA). Diffraction data were collected and processed with CrystalClear/d*Trek (17). The final dataset for PCNA:TIP was collected at the National Synchrotron Light Source, Brookhaven National Laboratory. Statistics for the data collection are shown in Table 1.
Table 1. Data collection and refinement statistics for PCNA:TIP and PCNA:FEN-1.
| PCNA withTIP (PDB ID: 5DA7) | PCNA with FEN-1 peptide (PDB ID: 5DAI) | |
|---|---|---|
| Diffraction data | ||
| space group | F222 | R3:H |
| cell dimensions (a,b,c) (Å) | 82.34, 184.08, 330.68 | 152.07, 152.07, 35.24 |
| resolution range (Å) | 30.0–2.8 | 28.8–2.0 |
| no. measured intensities | 514,688 | 66,808 |
| no. unique reflections | 30,707 (2,720)a | 20,143 (2,077) |
| average redundancy | 19.8 (6.3) | 3.31 (3.14) |
| % completeness | 98.9 (89.0) | 98.2 (97.9) |
| Rmerge | 0.116 (0.825) | 0.043 (0.419) |
| average I/sigI | 36.6 (2.2) | 11.9 (2.2) |
| Wilson B-factor | 80.6 | 43.7 |
| Refinement | ||
| resolution limits (Å) | 30.0–2.8 | 28.8–2.0 |
| R-factor (95% of the data) | 0.201 (0.300) | 0.182 (0.274) |
| Rfree (5% of the data) | 0.261 (0.359) | 0.224 (0.331) |
| rmsd bond lengths (Å) | 0.010 | 0.008 |
| rmsd bond angles (º) | 1.36 | 1.14 |
| Ramachandran favored (%) | 93.4 | 97.7 |
| Ramachandran outliers (%) | 1.8 | 0.0 |
| no. polypeptide chains in asymmetric unit | 2 PCNA, 2 TIP proteins | 1 PCNA, 1 FEN-1 peptide |
aNumbers in parentheses denote the values for the highest resolution shell.
Structure determination
Structures were determined by molecular replacement using the program PHASER (18). The search structure for PCNA:TIP and for PCNA:FEN-1 was the structure of the single polypeptide chain of PCNA [protein data base (PDB) ID: 3LX1]. In the PCNA:FEN-1 structure, FEN-1 peptide residues 330–338 are seen in the electron density. The TIP protein and FEN-1 peptide were built using the graphics program COOT (19). The resulting models were adjusted using COOT and refined against the electron density using REFMAC5 (20,21) and PHENIX (22–24). The final refinement statistics are summarized in Table 1. PyMOL (The PyMOL Molecular Graphics System. DeLano Scientific, San Carlos, CA, USA) or Chimera (25) was used to generate the molecular figures. In the PCNA:TIP structure, residues 1–27 and 63–64 of the TIP protein are not seen in the electron density. There is some unmodeled density between crystallographically related copies of chain A residues 161 and 187.
The initial electron density map for the PCNA:TIP crystals posed difficulty in fitting initially, using the PCNA monomer from PDB ID: 3LX1 as the search model, because the relationship of the two domains of PCNA is altered in the PCNA:TIP structure. After the complete PCNA protein was fit properly into the density map, there was still obvious density in the PIP motif binding site. Examination of the TIP sequence shows that there are two eight-residue stretches that satisfy a non-canonical PIP motif (vide infra), one starting at residue 2 (DRKLDEFI) and one starting at residue 31 (PTSLDSFL). Since there was electron density for several residues N-terminal to the first residue, the PIP motif sequence was built starting with residue 31 of TIP, not residue 2. Although there is not density for all of the side chains of TIP, there is sufficient resolution to confirm TIP residue 31 as the starting position. After the PIP motif residues were built, it became clear that there was considerable density beyond the C-terminal end of the non-canonical PIP sequence in TIP, and that this additional density represented a helix. Assignment of the TIP residues bound to PCNA starting at residue 31 instead of residue 2 was confirmed by solving a crystal structure using the TIP peptide (residues 28–64) and PCNA. PCNA:TIP and PCNA:TIP peptide crystallize in the same space group and have 72% solvent content. The PCNA in the PCNA:TIP peptide model was refined to a crystallographic R factor of 0.335 (R free 0.402) before the peptide model was added. When the peptide was added, the R factor dropped to 0.285 (R free 0.337). The core root-mean-square (rms) between the structure with TIP protein and the structure with the peptide is 0.3 Å. The presence of the peptide in the same position as the bound portion of TIP confirms the assignment of the sequence in the PCNA:TIP structure. Since the PCNA:TIP peptide dataset is at lower resolution (3.4 Å), the remainder of this paper will refer to the structure with the full length TIP protein. The PISA program (http://www.ebi.ac.uk/pdbe/prot_int/pistart.html) is designed to assist in defining protein interfaces, surfaces and assemblies (26) and was used in structural analysis.
Analysis of PCNA and PCNA complexes in solution
Solution studies with PCNA:TIP used the N-terminally truncated TIP peptide (residues 28–64) which has improved solubility at 100–200 mM NaCl over the full length TIP protein which requires 0.5 M NaCl. PCNA was dialyzed into Tris buffer containing 50 mM Tris–HCl (pH 7.3), 200 mM NaCl at 35–70 μM protein concentration using a 10 kDa molecular weight cut off (MWCO) membrane. TIP or FEN-1 peptides were added to PCNA in a PCNA:peptide ratio of 1:1 or 1:2. For SAXS experiments, the PCNA:TIP peptide complex was concentrated in an Amicon stirred cell with a 1 kDa MWCO membrane. A variant form of PCNA that was engineered to be monomeric (mPCNA) (16) was used as a reference for a PCNA monomer. The mPCNA protein was concentrated using an Amicon 0.5 concentrator with 10 kDa MWCO, and protein concentration was determined by absorbance at 280 nm using an extinction coefficient of 17,780 M−1cm−1. PCNA:TIP does not contain Trp residues and was difficult to quantitate. We determined an extinction coefficient of 7,400 M−1cm−1 at 280 nm for PCNA:TIP after quantitation from amino acid analysis (UC Davis Molecular Structure Facility). SAXS data were collected for the PCNA:TIP peptide complex at 6.4, 4.8, 4.1 and 2.1 mg/ml, a complex of PCNA:FEN-1 peptide at 7.3 and 5.5 mg/ml and the monomeric PCNA mutant at 1.8 mg/ml. The PCNA proteins and PCNA:peptide complexes were purified to a single peak using size exclusion chromatography in Tris buffer containing 50 mM Tris–HCl (pH 7.3), 200 mM NaCl, prior to SAXS data collection (Supplementary Experimental Procedures and Supplementary Table S1) and dialyzed overnight against Tris buffer. The buffer outside of the dialysis bag was used as a background measurement for SAXS. Sample dilutions were made using matching buffers from the dialysate and SAXS data were collected on the original concentration as well as the dilutions.
Small-angle X-ray scattering
SAXS data were collected at the Advanced Photon Source beamline 12-ID-B of Argonne National Laboratories, over the range 0.003 < q < 0.665 Å−1 where q = (4π/λ)sinθ and 2θ is the scattering angle. The energy of the X-ray beam was 18 keV and the sample to Pilatus 2M detector distance was 3 m. Thirty two-dimensional images were collected for each buffer and sample using a flow cell with an exposure time of 0.5–1.0 s at room temperature, and were reduced to one-dimensional scattering profiles using Matlab (The MathWorks Inc., Natick, MA, 2000, USA). No radiation damage was observed, as determined by comparing the intensities from sequentially acquired images. Primus (27) was used for Guinier analysis after background subtraction to determine the radius of gyration (Rg) and extrapolated intensity at zero scattering angle (Io) (Supplementary Figure S2 and Supplementary Table S2). Data were collected at several concentrations.
Experimental SAXS data were fitted to structural models using AXES formalism (28). For mPCNA and the PCNA:TIP complex, AXES was modified to include a two-member structural ensemble with weight optimization [reviewed in (29)]. The discrepancy between the modeled and experimental data is described by χ values. The coordinates used for the structural models of PCNA:FEN-1 were from the X-ray crystal coordinates (this work), and for free PCNA from the X-ray crystal structure (PDB ID: 3LX1). For mPCNA, a subset of coordinates from PDB ID: 3LX2 were used including (i) subunit A from PDB ID: 3XL2 as a monomer model and (ii) subunits A and B from PDB ID: 3LX2 as a model for a partial ring dimer. Model coordinates for PCNA:TIP were taken from (i) one of the PCNA:TIP complexes from the asymmetric unit for a 1:1 complex (Supplementary Figure S3A), (ii) both complexes in the asymmetric unit that create a 2:2 end-to-end linear structure (Supplementary Figure S3C) and (iii) two PCNA:TIP molecules (2:2) from neighboring unit cells that have PCNA tail-to-tail β strand contacts in a partial ring structure (Supplementary Figure S3B).
RESULTS
PCNA-TIP structure
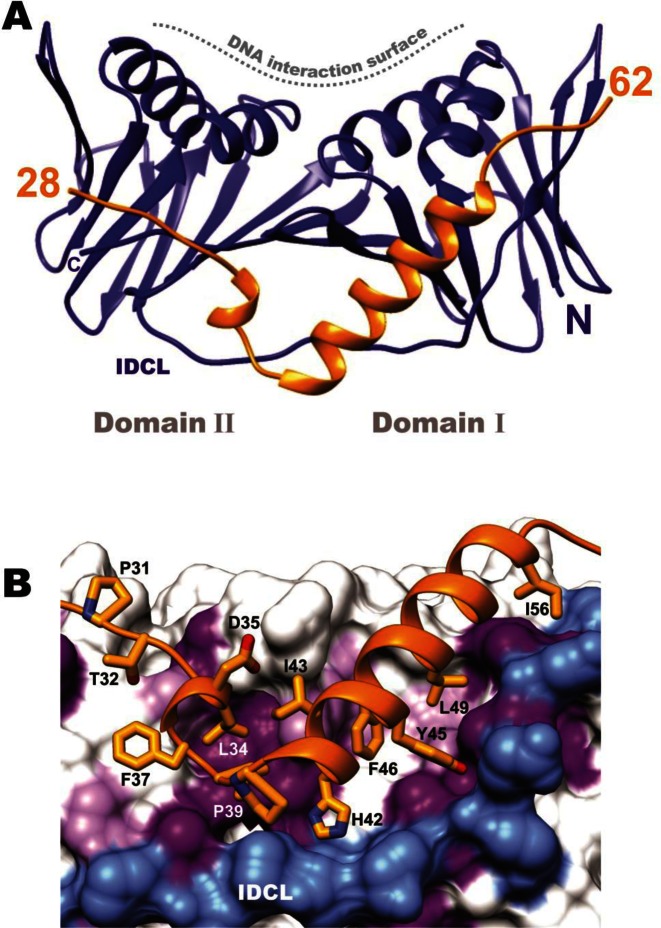
The 3D structure of TIP bound to PCNA was solved by X-ray crystallography to 2.8 Å resolution (Figure 2, Table 1). The monomer of PCNA in the complex maintains an overall similar structure to free PCNA that is comprised of two similar domains (domain I, residues 2–116 and domain II, residues 131–247), each containing two α-helices and two β-sheets which are connected by the IDCL (12). The rms deviation between the unligated PCNA polypeptide from the trimer and PCNA in the TIP complex is 2.2 Å overall and 1.0 Å and 0.8 Å between domains I and II, respectively. TIP binds on the outside of PCNA, away from the helices that surround DNA in the PCNA trimer (Figure 2A). TIP residues 1–27 are not observed in the crystal due to disorder; however, residues 28–62 are observed and form a distinct structure. TIP residues 31–38 bind at the PIP site on PCNA, which is surprising, since TIP does not contain a canonical PIP motif (QXXhXXaa, described above). There are numerous contacts between TIP residues at the PIP site and PCNA that include hydrophobic interactions, ionic interactions and hydrogen bonds. The amino acid sequence of TIP that follows the PIP binding region forms a turn and then continues as a helix across the β-sheet of domain I (Figure 2).
Figure 2.
The structure of PCNA:TIP complex. (A) Ribbon diagram of PCNA (blue) bound to the TIP protein (gold) (PDB ID: 5DA7), showing the short helix in the PIP binding pocket followed by a turn and a helix that binds across the β-sheet of PCNA Domain I. TIP does not interact with the helices on PCNA that form the interior of the DNA clamp, located at the top in the figure. (B) Close up of the PCNA:TIP interactions; TIP is colored gold and residues interacting with PCNA are labeled. Hydrophobic residues on the surface of PCNA are shaded purple. The IDCL is shown in blue, for reference.
PCNA:FEN-1 structure
In order to compare the TIP interactions at the PIP site on PCNA, the structure of T. kodakarensis FEN-1 peptide with PCNA was determined by X-ray crystallography to 2.0 Å resolution (Table 1, Supplementary Figure S4). FEN-1 is one of the proteins that interact with and is regulated by PCNA and in all species studied it contains a canonical PIP motif (30,31). The structure of T. kodakarensis FEN-1 bound to PCNA is very similar to PCNA:FEN-1 structures from other organisms (32–35).
Non-canonical PIP motif in TIP
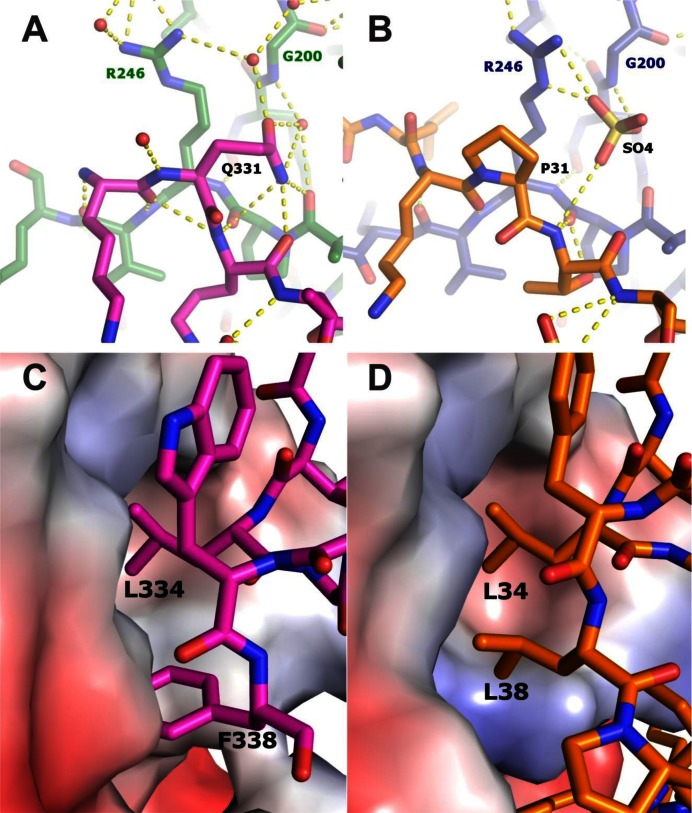
The structure of TIP in the PIP binding site is very similar to other PIP peptides and to the FEN-1 structure determined here. Figure 1B shows a comparison of the PIP site structure of TIP in the PCNA:TIP complex superimposed with other known PIP peptides. It appears that TIP does form a PIP site structure, although with a non-canonical sequence. Important residues for PIP site binding are a hydrophobic fourth residue and two hydrophobic aromatic residues, as well as the initial glutamine. TIP contains two canonical PIP site residues, L34 and F38, that have very similar interactions with PCNA as FEN-1 peptide residues L334 and W338 (Supplementary Figure S5). FEN-1 and TIP also contain an acidic residue (E335 and D35, respectively) in the PIP site that forms a hydrogen bond to the side chain of PCNA R45. Differences in PIP site interactions between FEN-1 and TIP structures occur for the non-canonical residues of TIP. Specifically, P31 is the first residue in the PIP site, but it does not enter the ‘Q’ pocket on PCNA. In the PCNA:TIP structure, there is a sulfate ion in the ‘Q’ pocket that forms hydrogen bonds to the amide group of G200 and the side chain Nϵ and Nη2 atoms of R246 (Figure 3B). These contacts mimic Q331 contacts in the PCNA:FEN-1 structure (Figure 3A). The final residue of TIP in the PIP site is L38. Although TIP does not contain a second aromatic residue, this leucine conserves the hydrophobic interaction (Figure 3C and D).
Figure 3.
Comparison of TIP and FEN-1 interactions with PCNA. (A) Local contacts for glutamine in position 331 of the PIP motif of FEN-1 (first residue of the PIP-motif). The pink color shows the PIP motif residues and the green color shows the PCNA residues. The small red balls represent water molecules and the yellow dashed lines represent hydrogen bonds. (B) Local contacts for the non-canonical proline in position 31 of the PIP motif of TIP (first residue of the PIP-motif). TIP is in gold and PCNA is in blue. Panels C and D, hydrophobic pocket for residue 8 of the PIP motifs. (C) F338 of FEN-1 is in a hydrophobic pocket shared with L334. (D) TIP residues L38 and L34 are in the same hydrophobic pocket.
Interaction of the C-terminal helix of TIP with PCNA
The extensive contacts of the TIP helix to PCNA is reflected in analysis of the interface area of both complexes, which is 2.5 times larger in the TIP complex (Supplementary Table S3). The 15 residue helix of TIP extends from H42 to I56. It binds in a path created by movement of the IDCL that exposes hydrophobic residues on PCNA and creates a pocket for TIP residues I43, F46, L49 and I56 (Figure 2B). There are several hydrogen bonds between TIP and PCNA as well, between TIP residues R50 and S53 near the C-terminal end of the helix to E26 and N72 of PCNA and also between the side chain hydroxyl of TIP Y45 to the backbone carbonyl of PCNA E119. The extension of the 17 residues past the PIP motif of TIP interacts with PCNA in a manner that is in sharp contrast to the structures of human PCNA with FEN-1 and p21 (PDB IDs: 1U7B and 1AXC, respectively). In these structures, the sequence that extends beyond the PIP motif forms β-strand connections to the IDCL, but does not perturb the PCNA ring structure.
Conformational changes of PCNA in the PCNA:TIP complex
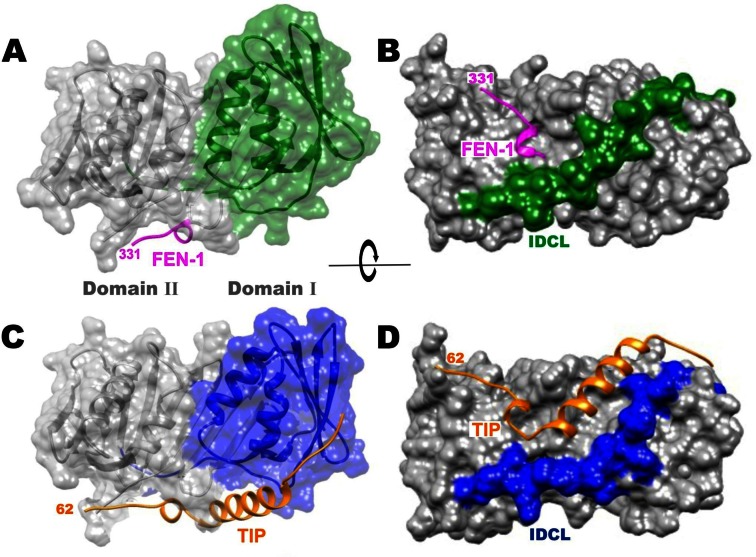
The binding pocket for the TIP helix is formed by >7 Å shift of the IDCL (Figure 4B and D). Movement of the IDCL is a plausible adjustment, since it has been shown to be flexible in other structures [(36) reviewed in (10)]. Further structural changes are evident between domains I and II of PCNA:TIP (Figure 4). These can be visualized by comparing the surface of domain I between the PCNA:FEN-1 structure (Figure 4A, green) with that in the PCNA:TIP structure (Figure 4C, blue), where domain II is held in the same position in both images. The domains move away from each other, such that the interface between them is reduced by 100 Å2 (Supplementary Table S3). Additionally, the helices of domain I are tilted toward the right, and much of the domain also rotates slightly toward the back of the image. The overall effect is a twist between domains I and II in PCNA:TIP. It should be emphasized that domains I and II remain very similar between the two structures (Table 2). It is largely the orientation of the two domains relative to one another in the PCNA:TIP structure that is different. The β-sheets are intact for both domains in this structure but the curvature of the β-sheet in domain II is increased to accommodate the twist.
Figure 4.
TIP binding to PCNA results in a large movement of the IDCL. Panels (A) and (B), PCNA:FEN-1 structure with FEN-1 in magenta. Panels (C) and (D), PCNA:TIP structure with TIP in gold. In panels (A) and (C) PCNA domain II is in the same orientation while changes in domain I are highlighted in color (green and blue). Panels (B) and (D) are a rotated view of panels (A) and (C) to highlight the 7 Å movement of the IDCL (green and blue) between the two structures.
Table 2. Structural comparison of PCNA and PCNA complexes.
| Structural comparisonsa | PCNA:TIP rms (Å) | PCNA:FEN-1 rms (Å) |
|---|---|---|
| Overall | 2.2 (245 residues) | 1.1 (246 residues) |
| Domain I (residues 2–116) | 1.0 | 0.8 |
| Domain II (residues 131–247) | 0.8 | 0.4 |
aThe domains were compared in COOT to a monomer of free PCNA (PDB ID: 3LX1).
Structural changes in PCNA:TIP prevent trimer formation
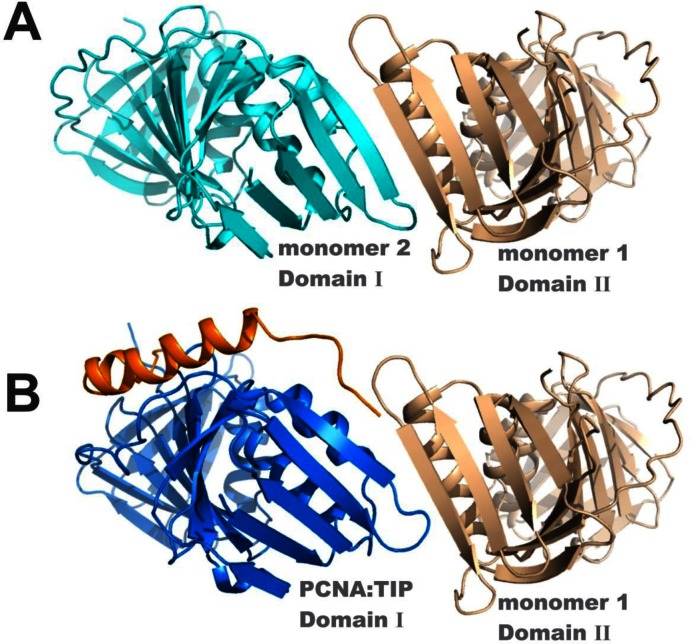
A result of these conformational changes in PCNA:TIP is that the head-to-tail arrangement of the trimer interface is no longer possible. Figure 5A shows the trimer interface contacts in free PCNA as an antiparallel β-strand between domain I of one monomer and domain II of another monomer that form an extended β-sheet across both monomers (cyan to tan) in the trimer form. In Figure 5B, domain II of PCNA:TIP is superimposed with domain II of monomer 2, and then monomer 2 was removed, so that the relative position of PCNA:TIP domain I is apparent. PCNA:TIP cannot form a trimer because the β-strands can no longer line up to form the necessary intermolecular contacts (blue to tan) to make stable intermolecular interactions.
Figure 5.
TIP binding breaks the interface between monomers of PCNA. (A) The interface between monomers of the trimer ring in free PCNA involves a smooth continuation of the β-sheets (cyan to tan). (B) The twist of domain I in the PCNA:TIP structure prevents trimer formation. To create panel B, the domain II of PCNA:TIP (blue:gold) has been superimposed onto the domain II of one of the monomers in the free PCNA trimer (cyan), then that free PCNA domain was removed from the graphic. The β-strands (from blue to tan) no longer line up to form a hydrogen bonded continuous sheet. The C-terminal end of TIP is fairly flexible since residues 57–61 have higher average B-factors.
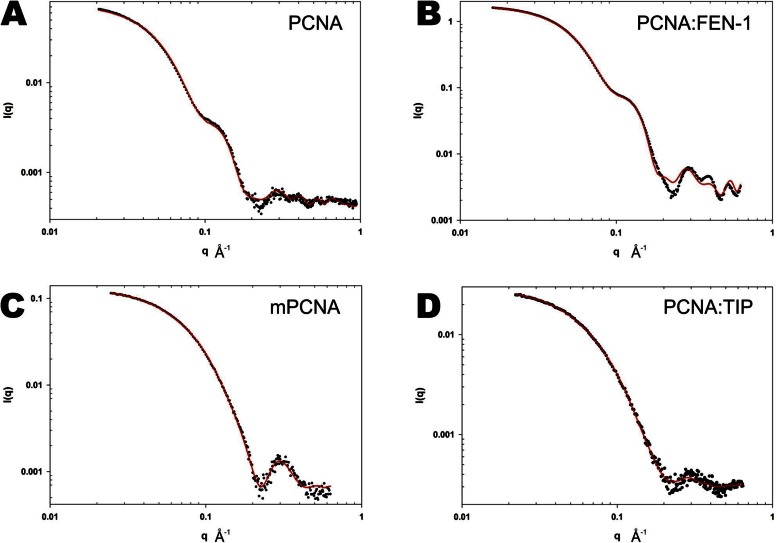
SAXS analysis shows disruption of the PCNA trimer by TIP
SAXS can readily distinguish between different shapes and sizes of proteins. In particular, the hollow ring structure of a PCNA trimer will produce a different scattering curve and invariants from Guinier analysis (Io, Rg) than that of a globular structure, such as a 1:1 complex of PCNA:TIP. SAXS data were collected for four different structures: free PCNA, PCNA:FEN-1, mPCNA and PCNA:TIP. The experimental SAXS data for free PCNA is shown in Figure 6A and for PCNA:FEN-1 in Figure 6B (data points). Inflections in these scattering curves are indicative of a hollow ring structure (37). For PCNA:FEN-1, the SAXS data were fit to the X-ray structure determined here (Figure 6B, solid line, χ = 2.4). For free PCNA, the SAXS data were fit to the X-ray crystal structure (PDB ID: 3LX1) (Figure 6A, solid line, χ = 0.44). T. kodakarensis PCNA has been observed to be a less stable trimer than other PCNAs (12,13). To address the possibility of a partially dissociated PCNA protein, alternative fits to the experimental SAXS data were made using equilibrium models of a monomer-trimer and also of a monomer-dimer (partial ring). Both equilibrium models resulted in poor fits to the experimental SAXS data. Therefore, PCNA retains its trimeric structure in the experiments reported here (Supplementary Table S1 and Figure 6). The crystal structure trimer models of PCNA and PCNA:FEN-1 fit the experimental SAXS data well at low q, with some differences at q-values > 0.16. These differences may be due to variation between the crystal lattice and solution states of PCNA and PCNA:FEN-1. The experimental SAXS data for mPCNA is shown in Figure 6C (data points). The best fit of the experimental SAXS data to a structure was obtained using a monomer-dimer equilibrium structure model (χ ∼ 0.35, solid line, Figure 6C). This fit indicates the mPCNA protein is largely, but not completely, monomeric in solution and contains a small population of dimer (∼12%).
Figure 6.
Small angle X-ray scattering data of PCNA and PCNA complexes in solution. In each of the graphs, the experimental data is shown as data points and the fit to the data using models from the X-ray structures calculated with AXES is displayed as a solid red line. (A) PCNA trimer, (B) PCNA:FEN-1 trimer, (C) mPCNA and (D) PCNA:TIP. Both the mPCNA and PCNA:TIP data fit better to a two-state model than to a single structure.
The experimental SAXS data for PCNA:TIP peptide is shown in Figure 6D (data points). By comparing the SAXS data for PCNA:TIP with trimeric PCNA (Figure 6A and B) and monomeric PCNA (mPCNA, Figure 6C), it is clear that PCNA:TIP is not a trimeric ring and is thus unable to function as a DNA clamp. A fit of the SAXS data to the crystal structure of PCNA:TIP in a 1:1 complex (Model A, Supplementary Figure S3A) showed poor agreement. It appears that PCNA:TIP is in equilibrium between this state and a 2:2 dimer form. As noted in Table 1, there are two copies of PCNA:TIP complex in the asymmetric unit (Model C, Supplementary Figure S3C). There are also crystallographic contacts between PCNA molecules of neighboring unit cells (Model B, Supplementary Figure S3B). The protein contacts between unit cells consist of a β-strand in domain II (PCNA residues 174–180) that is anti-parallel to the same strand in domain II on another PCNA:TIP molecule. These tail-to-tail contacts are different from head-to-tail interactions in the free PCNA trimer. The fit of the experimental SAXS data representing an equilibrium state between Models A and C is shown as a solid line in Figure 6D. From this fit, the PCNA:TIP complex is 69% Model A and 31% Model C (χ ∼ 0.24) at 2 mg/ml protein concentration. The fit to the experimental SAXS data for an equilibrium state between Models A and B did not fit the experimental data as well (χ ∼ 0.41, Supplementary Figure S6). SAXS data collected at higher concentrations show an increase in self-association of PCNA:TIP, and the KD could be estimated at ∼100 μM. Therefore, PCNA:TIP appears to self-associate in a form similar to Model C that becomes largely 1:1 (Model A) at concentrations below 2 mg/ml.
DISCUSSION
Biochemical studies demonstrated that TIP binding to PCNA inhibits PCNA-dependent activities, and size exclusion chromatography indicated the formation of a smaller complex (16). With the X-ray structures and SAXS data presented here, TIP protein binding to PCNA results in trimeric ring dissociation. To date, most activities for PCNA require PCNA to encircle DNA. Thus, it is the disruption of the PCNA ring by TIP that leads to inhibition of PCNA-dependent enzymatic activities, as PCNA can no longer encircle the DNA.
Transient PCNA ring opening plays an important role during DNA replication in archaea and eukarya. During DNA replication, PCNA needs to be assembled around the primers on the lagging strand at the beginning of each Okazaki fragment (38). This activity is performed by the clamp loader complex, replication factor C (RFC), that opens the PCNA ring for assembly around the primers. In eukarya, in addition to RFC, there is an RFC-like complex (RLC) that contains four of the five RFC subunits, but the fifth one is replaced by Elg1. The Elg1-RLC role is to unload PCNA following the maturation of the Okazaki fragments (39). These unloaded PCNAs can be used for subsequent reloading by RFC. However, the function of TIP is different than either of these two complexes, as its role is to dissociate PCNA and thus block PCNA associated activities.
During PCNA loading and unloading from DNA, the interfaces between the PCNA subunits are open. It is not clear if in vivo TIP binding dissociates the intact PCNA ring or whether it interacts with opened or partially assembled rings generated by RFC or an archaeal enzyme with Elg1-RLC-like activity. In addition, it was previously shown that the T. kodakarensis PCNA does not form a stable trimeric ring in vitro (12–14). The oligomeric structure of the protein in vivo, however, is not clear and both trimeric (13) and monomeric (14) forms have been suggested. Under all solution measurements made in this study, PCNA was trimeric. Therefore, TIP can dissociate an intact trimeric PCNA ring. TIP also binds to monomeric PCNA (16), which suggests TIP would inhibit either the closed or open ring forms of PCNA.
In general, the stability of archaeal PCNA proteins is different from eukaryotic PCNA proteins and the bacterial sliding clamp, the β-subunits. Both eukaryotic PCNA proteins and β-subunits are very stable rings (40) and are only opened in an energy-dependent manner via ATP hydrolysis by the clamp loaders (RFC and the τ-complex, respectively) or the clamp unloader (Elg1-RLC). The data presented here show that the T. kodakarensis PCNA can be opened in an ATP-independent manner by the TIP protein. In addition, biochemical studies have shown that several archaeal PCNA proteins can assemble around DNA in a RFC-independent manner [for examples see (12,41–43)]. It would be of interest to see whether eukaryal and bacterial sliding clamps can also be opened in an ATP-independent manner and if proteins similar to TIP also exist in these domains.
The role of TIP in T. kodakarensis is not yet known. It is not clear at what stage of the cell cycle TIP is expressed or if it is expressed under specific circumstances (i.e. DNA damage) which require a halt in DNA replication. Future studies may address these questions. Nevertheless, the disruption of a DNA clamp by a small protein, like TIP, represents a new mechanism for PCNA regulation. It contrasts with previously described inhibitors of eukaryotic PCNA, such as the cell cycle regulator p21, that function as competitive inhibitors at the PIP site (44).
Supplementary Material
Acknowledgments
X-ray crystal data for this study were measured at beamline X29 of the National Synchrotron Light Source. For the SAXS experiments, we gratefully acknowledge use of SAXS core facility of Center for Cancer Research, National Cancer Institute (NCI). The shared scattering beamline 12-ID-B resource is allocated under the PUP-77 agreement between Yun-Xing Wang at the National Cancer Institute and Argonne National Lab (ANL). We thank Lixin Fan (NCI), Xiaobing Zuo (ANL) and Alex Grishaev for their expert support, and Sandra Gutierrez for help in the early stages of this study. We also would like to thank Lori Kelman and the anonymous reviewers for their helpful comments on the manuscript.
Certain commercial equipment, instruments and materials are identified in this paper in order to specify the experimental procedure. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the material or equipment identified is necessarily the best available for the purpose.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of Standards and Technology.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kelman Z., O'Donnell M. Structural and functional similarities of prokaryotic and eukaryotic DNA polymerase sliding clamps. Nucleic Acids Res. 1995;23:3613–3620. doi: 10.1093/nar/23.18.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jeruzalmi D., O'Donnell M., Kuriyan J. Clamp loaders and sliding clamps. Curr. Opin. Struct. Biol. 2002;12:217–224. doi: 10.1016/s0959-440x(02)00313-5. [DOI] [PubMed] [Google Scholar]
- 3.Kelman L.M., Kelman Z. Archaeal DNA replication. Annu. Rev. Genet. 2014;48:71–97. doi: 10.1146/annurev-genet-120213-092148. [DOI] [PubMed] [Google Scholar]
- 4.MacNeill S.A. PCNA binding proteins in the archaea: novel functionality beyond the conserved core. Curr. Genet. 2016 doi: 10.1007/s00294-016-0577-3. doi:10.1007/s00294-016-0577-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moldovan G.L., Pfander B., Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Vivona J.B., Kelman Z. The diverse spectrum of sliding clamp interacting proteins. FEBS Lett. 2003;546:167–172. doi: 10.1016/s0014-5793(03)00622-7. [DOI] [PubMed] [Google Scholar]
- 7.Mailand N., Gibbs-Seymour I., Bekker-Jensen S. Regulation of PCNA-protein interactions for genome stability. Nat. Rev. Mol. Cell Biol. 2013;14:269–282. doi: 10.1038/nrm3562. [DOI] [PubMed] [Google Scholar]
- 8.Warbrick E. PCNA binding through a conserved motif. Bioessays. 1998;20:195–199. doi: 10.1002/(SICI)1521-1878(199803)20:3<195::AID-BIES2>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 9.Maga G., Hubscher U. Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J. Cell Sci. 2003;116:3051–3060. doi: 10.1242/jcs.00653. [DOI] [PubMed] [Google Scholar]
- 10.Dieckman L.M., Freudenthal B.D., Washington M.T. PCNA structure and function: insights from structures of PCNA complexes and post-translationally modified PCNA. Subcell. Biochem. 2012;62:281–299. doi: 10.1007/978-94-007-4572-8_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan M., Kelman L.M., Kelman Z. The archaeal PCNA proteins. Biochem. Soc. Trans. 2011;39:20–24. doi: 10.1042/BST0390020. [DOI] [PubMed] [Google Scholar]
- 12.Ladner J.E., Pan M., Hurwitz J., Kelman Z. Crystal structures of two active proliferating cell nuclear antigens (PCNAs) encoded by Thermococcus kodakaraensis. Proc. Natl. Acad. Sci. U.S.A. 2011;108:2711–2716. doi: 10.1073/pnas.1019179108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan M., Santangelo T.J., Čuboňová L., Li Z., Metangmo H., Ladner J., Hurwitz J., Reeve J.N., Kelman Z. Thermococcus kodakarensis has two functional PCNA homologues but only one is required for viability. Extremophiles. 2013;17:453–461. doi: 10.1007/s00792-013-0526-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuba Y., Ishino S., Yamagami T., Tokuhara M., Kanai T., Fujikane R., Daiyasu H., Atomi H., Ishino Y. Comparative analyses of the two proliferating cell nuclear antigens from the hyperthermophilic archaeon, Thermococcus kodakarensis. Genes Cells. 2012;17:923–937. doi: 10.1111/gtc.12007. [DOI] [PubMed] [Google Scholar]
- 15.Li Z., Santangelo T.J., Čuboňová L., Reeve J.N., Kelman Z. Affinity purification of an archaeal DNA replication protein network. MBio. 2010;1:e00221–e00210. doi: 10.1128/mBio.00221-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z., Huang R.Y., Yopp D.C., Hileman T.H., Santangelo T.J., Hurwitz J., Hudgens J.W., Kelman Z. A novel mechanism for regulating the activity of proliferating cell nuclear antigen by a small protein. Nucleic Acids Res. 2014;42:5776–5789. doi: 10.1093/nar/gku239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pflugrath J.W. The finer things in X-ray diffraction data collection. Acta Crystallogr. D Biol. Crystallogr. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 18.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 20.Collaborative Computational Project, N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 21.Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 22.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen V.B., Arendall W.B., 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., Terwilliger T.C., Urzhumtsev A., Zwart P.H., Adams P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. 2004. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 26.Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 27.Konarev P.V., Volkov V.V., Sokolova A.V., Koch M.H.J., Svergun D.I. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Cryst. 2003;36:1277–1282. [Google Scholar]
- 28.Grishaev A., Guo L., Irving T., Bax A. Improved fitting of solution X-ray scattering data to macromolecular structures and structural ensembles by explicit water modeling. J. Am. Chem. Soc. 2010;132:15484–15486. doi: 10.1021/ja106173n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tuukkanen A.T., Svergun D.I. Weak protein-ligand interactions studied by small-angle X-ray scattering. FEBS J. 2014;281:1974–1987. doi: 10.1111/febs.12772. [DOI] [PubMed] [Google Scholar]
- 30.Balakrishnan L., Bambara R.A. Flap Endonuclease 1. Annu. Rev. Biochem. 2013;82:119–138. doi: 10.1146/annurev-biochem-072511-122603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finger L.D., Atack J.M., Tsutakawa S., Classen S., Tainer J., Grasby J., Shen B. The wonders of flap endonucleases: structure, function, mechanism and regulation. Subcell. Biochem. 2012;62:301–326. doi: 10.1007/978-94-007-4572-8_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakurai S., Kitano K., Yamaguchi H., Hamada K., Okada K., Fukuda K., Uchida M., Ohtsuka E., Morioka H., Hakoshima T. Structural basis for recruitment of human flap endonuclease 1 to PCNA. EMBO J. 2005;24:683–693. doi: 10.1038/sj.emboj.7600519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dore A.S., Kilkenny M.L., Jones S.A., Oliver A.W., Roe S.M., Bell S.D., Pearl L.H. Structure of an archaeal PCNA1-PCNA2-FEN1 complex: elucidating PCNA subunit and client enzyme specificity. Nucleic Acids Res. 2006;34:4515–4526. doi: 10.1093/nar/gkl623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chapados B.R., Hosfield D.J., Han S., Qiu J., Yelent B., Shen B., Tainer J.A. Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell. 2004;116:39–50. doi: 10.1016/s0092-8674(03)01036-5. [DOI] [PubMed] [Google Scholar]
- 35.Bruning J.B., Shamoo Y. Structural and thermodynamic analysis of human PCNA with peptides derived from DNA polymerase-δ p66 subunit and flap endonuclease-1. Structure. 2004;12:2209–2219. doi: 10.1016/j.str.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 36.De Biasio A., Campos-Olivas R., Sanchez R., Lopez-Alonso J.P., Pantoja-Uceda D., Merino N., Villate M., Martin-Garcia J.M., Castillo F., Luque I., et al. Proliferating cell nuclear antigen (PCNA) interactions in solution studied by NMR. PLoS One. 2012;7:e48390. doi: 10.1371/journal.pone.0048390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Putnam C.D., Hammel M., Hura G.L., Tainer J.A. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q. Rev. Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 38.Yao N.Y., O'Donnell M. The RFC clamp loader: structure and function. Subcell. Biochem. 2012;62:259–279. doi: 10.1007/978-94-007-4572-8_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kubota T., Katou Y., Nakato R., Shirahige K., Donaldson A.D. Replication-coupled PCNA unloading by the Elg1 complex occurs genome-wide and requires Okazaki fragment ligation. Cell Rep. 2015;12:774–787. doi: 10.1016/j.celrep.2015.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yao N., Turner J., Kelman Z., Stukenberg P.T., Dean F., Shechter D., Pan Z.Q., Hurwitz J., O'Donnell M. Clamp loading, unloading and intrinsic stability of the PCNA, β and gp45 sliding clamps of human, E. coli and T4 replicases. Genes Cells. 1996;1:101–113. doi: 10.1046/j.1365-2443.1996.07007.x. [DOI] [PubMed] [Google Scholar]
- 41.Kelman Z., Hurwitz J. A unique organization of the protein subunits of the DNA polymerase clamp loader in the archaeon Methanobacterium thermoautotrophicum ΔH. J. Biol. Chem. 2000;275:7327–7336. doi: 10.1074/jbc.275.10.7327. [DOI] [PubMed] [Google Scholar]
- 42.Matsumiya S., Ishino S., Ishino Y., Morikawa K. Physical interaction between proliferating cell nuclear antigen and replication factor C from Pyrococcus furiosus. Genes Cells. 2002;7:911–922. doi: 10.1046/j.1365-2443.2002.00572.x. [DOI] [PubMed] [Google Scholar]
- 43.Seybert A., Scott D.J., Scaife S., Singleton M.R., Wigley D.B. Biochemical characterisation of the clamp/clamp loader proteins from the euryarchaeon Archaeoglobus fulgidus. Nucleic Acids Res. 2002;30:4329–4338. doi: 10.1093/nar/gkf584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gibbs E., Kelman Z., Gulbis J.M., O'Donnell M., Kuriyan J., Burgers P.M.J., Hurwitz J. The influence of the proliferating cell nuclear antigen-interacting domain of p21CIP1 on DNA synthesis catalyzed by the human and Saccharomyces cerevisiae polymerase δ holoenzymes. J. Biol. Chem. 1997;272:2373–2381. doi: 10.1074/jbc.272.4.2373. [DOI] [PubMed] [Google Scholar]
- 45.Nishida H., Mayanagi K., Kiyonari S., Sato Y., Oyama T., Ishino Y., Morikawa K. Structural determinant for switching between the polymerase and exonuclease modes in the PCNA-replicative DNA polymerase complex. Proc. Natl. Acad. Sci. U.S.A. 2009;106:20693–20698. doi: 10.1073/pnas.0907780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.