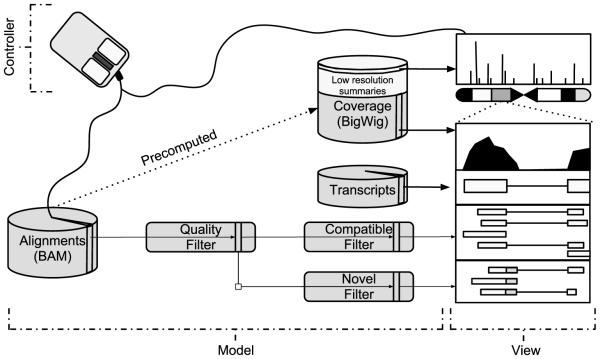
Fig. 4.
An application of the model–view–controller pattern and pre-computed summaries to genomic visualization. Coverage is displayed at two levels of resolution (whole chromosome and the current zoom) after efficient extraction from the multi-resolution BigWig file. The BAM file holding the read alignments is abstracted by a multi-stage data model, consisting of the BAM source, a dynamic read quality filter and two filters that effectively split the alignments according to compatibility with the known transcript annotations. The view contains several coordinately-zoomed plots, as well as an ideogram and coverage overview. Each plot obtains its data from one of the data model components. The controller might adjust the data model filter settings and the current zoom in response to user commands.