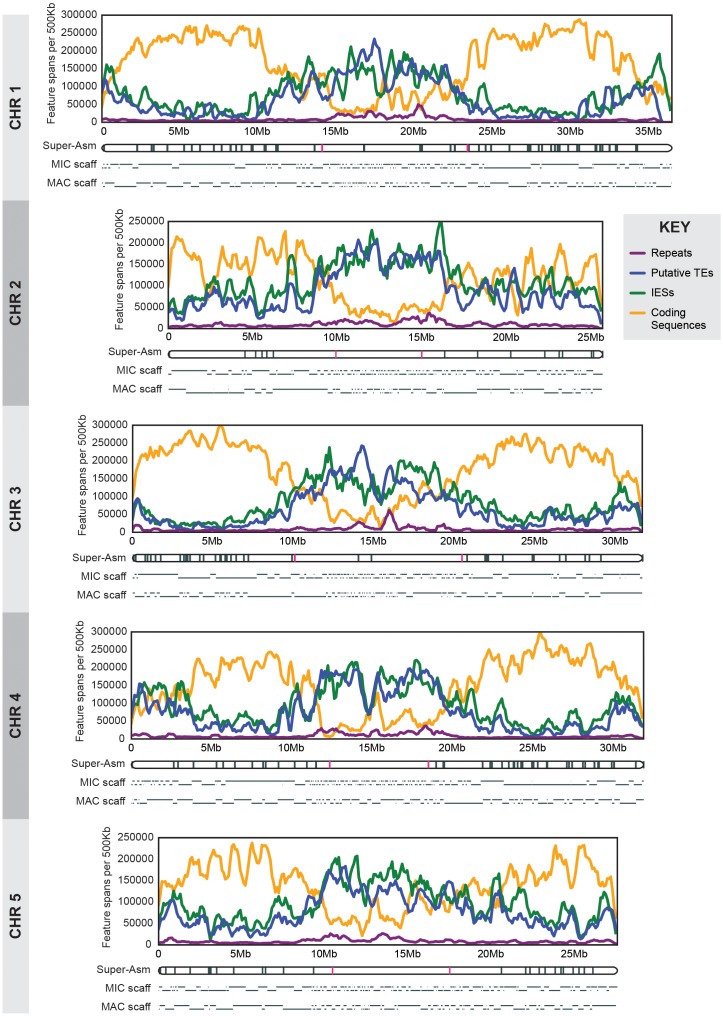
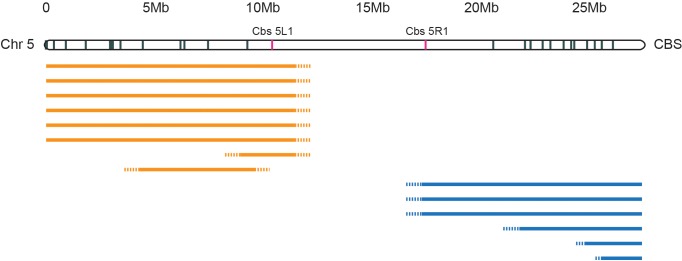
Figure 2. MIC chromosome landscapes.
For each chromosome, the top panel shows the density of several genomic features, measured as number of base pairs (span) per 500 kb sliding window (100 kb slide increment). Purple = simple sequence repetitive DNA (note that exclusion of those simple sequence repeats that overlap with TEs has minimal effect on the distribution pattern). Blue = putative TEs. Green = high-confidence IESs. Orange = protein-coding sequences. The corresponding chromosome-length super-assembly (Super-Asm) is shown immediately below, each Cbs indicated by a vertical tick. Red ticks indicate Cbs’s flanking putative centromeres (see main text and Figure 2—figure supplement 1). In the 'MIC-scaff' schematic, the scaffolds comprising each MIC chromosome super-assembly are depicted as horizontal lines (alternating in vertical position to delineate each from its neighbors). The ‘MAC-scaff’ schematic indicates the positions of MAC scaffolds (many of which are complete, fully sequenced MAC chromosomes) derived from the corresponding regions of the MIC chromosome. Note that, because IESs are absent from MAC scaffolds, their lengths are actually shorter, but for simplicity of viewing, these lengths have been stretched so that MAC-scaff endpoints line up with their corresponding positions in the MIC. Chromosomes are stacked so that their centers align vertically.