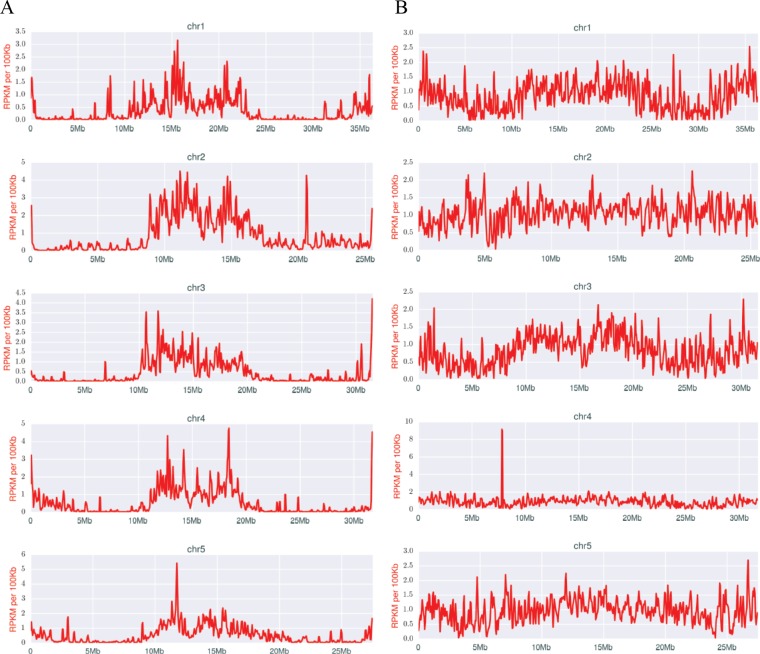
Figure 7. Densities of early (A) and late (B) scnRNAs on MIC chromosomes.
X-axis = position on MIC chromosome super-assembly; all graphs normalized to the same length. Early-scnRNAs were co-purified with Twi1p at three hpm and Late-scnRNAs with Twi11p at 10.5 hpm. Normalized numbers (Reads per kb per million reads [RPKM] in 50 kb bins) of sequenced 26–32-nt RNAs that uniquely map to the MIC genome are shown. A few locations on the chromosomal arms where Early- or Late-scnRNAs were extensively mapped (e.g. ~20.6 Mb on Chr2 for Early-scnRNA and ~7.8 Mb on Chr4 for Late-scnRNAs) were examined in detail, but we have failed to detect any obvious unusual sequence features at these loci to account for the observed enrichment.