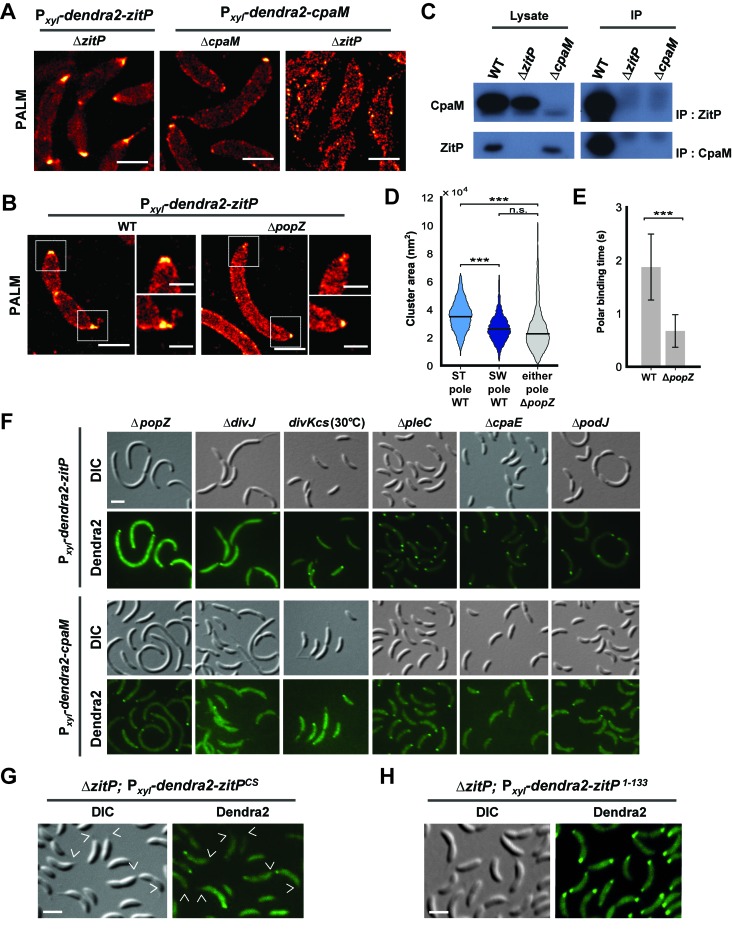
Figure 3. Distinct ZitP nanoscale assemblies and localization determinants.
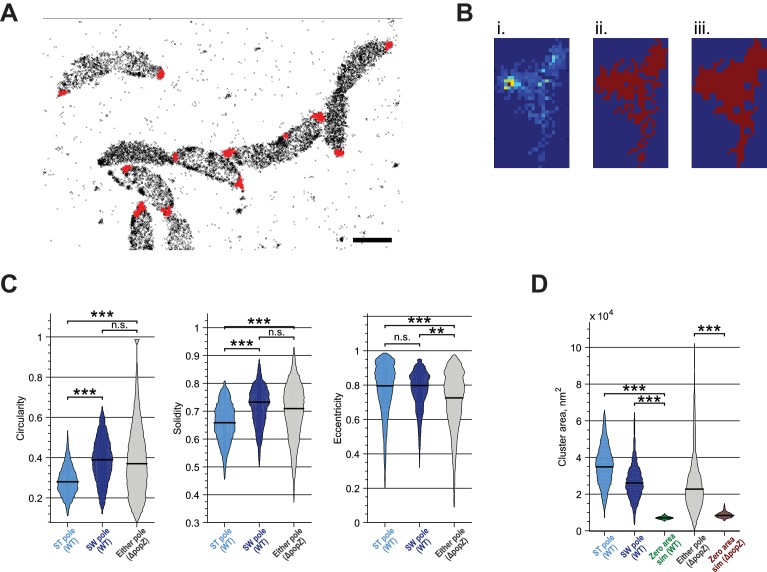
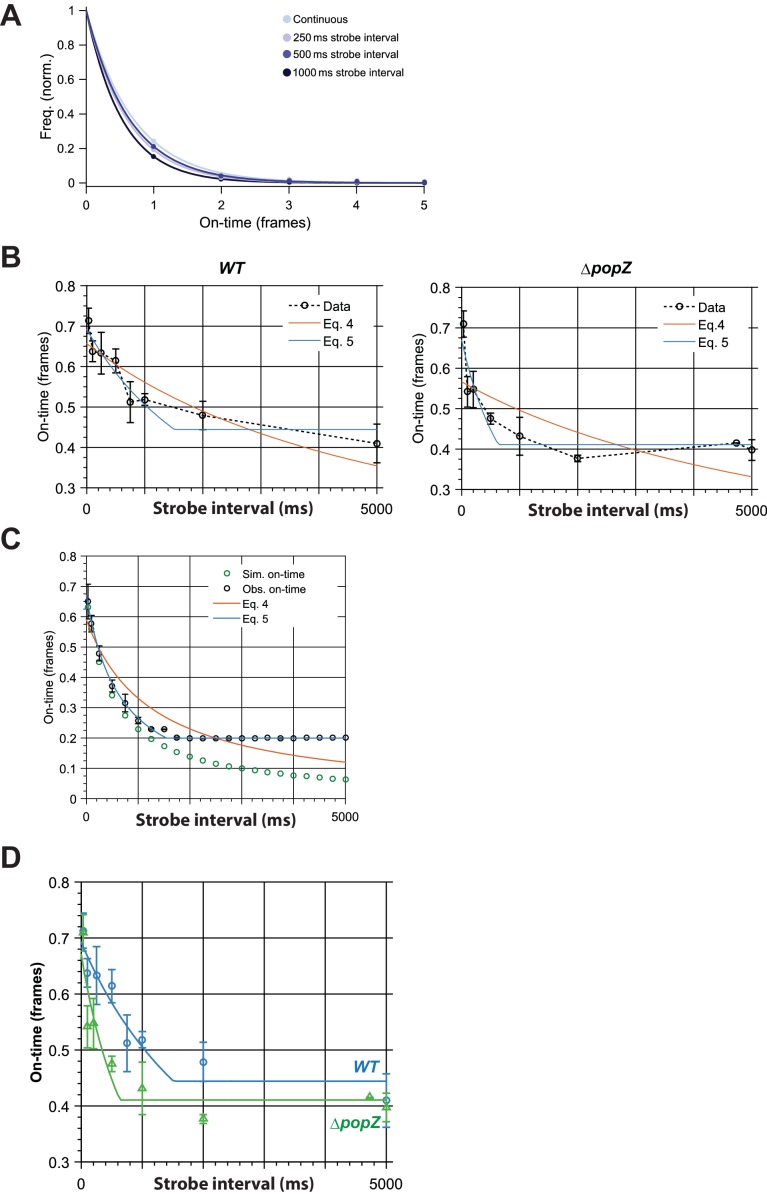
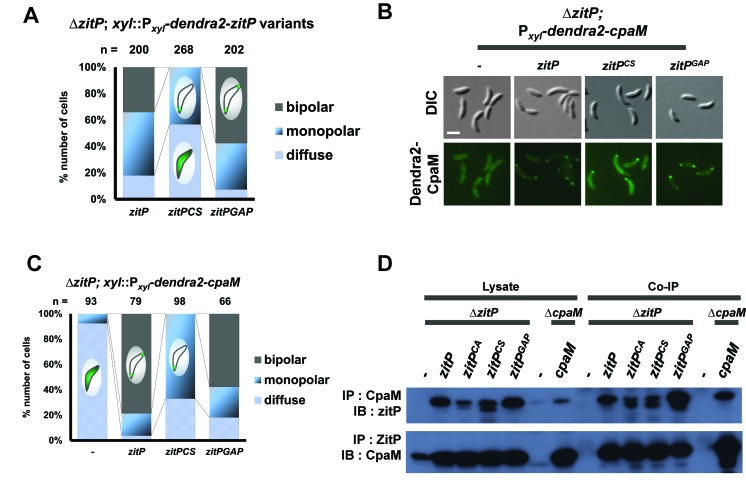
(A) Photo-activated light microscopy (PALM) imaging of Dendra2-ZitP or Dendra2-CpaM expressed from the xylose-inducible Pxyl promoter on a plasmid integrated at the chromosomal xylX locus in ΔzitP or ΔcpaM cells exposed to xylose 3 hours before imaging. Scale bar: 1 µm. (B) PALM imaging of Dendra2-ZitP in WT or ΔpopZ::Ω cells. We induced expression of Dendra2-ZitP from the xylose-inducible Pxyl promoter on a plasmid integrated at the chromosomal xylX locus by the addition of xylose 3 hours before imaging. Scale bar: 1 µm. Scale bar of zoomed images: 0.5 µm. (C) Co-immunoprecipitation (co-IP) of ZitP or CpaM with polyclonal antibodies to CpaM or ZitP, respectively. Immunoprecipitates and cell lysates from WT, ΔzitP or ΔcpaM cells were probed for the presence of ZitP or CpaM. (D) Projected area of the Dendra2-ZitP polar complex as determined by PALM from Dendra2-ZitP expressed in WT and ΔpopZ::Ω cells. Black lines indicate medians. Statistical significance from Mood’s median test: n.s, p>0.05; ***p<0.001. (E) ZitP polar binding times in WT and ΔpopZ::Ω cells, measured via single particle tracking PALM. Error bars indicate 95% confidence interval of the fit to the data (Figure 3—figure supplement 6D). Statistical significance from a 2 sample t-test: ***p=p<0.001. (F) Epifluorescence (Dendra2) and Nomarski (DIC) images depicting the localization of Dendra2-ZitP or Dendra2-CpaM in ΔpopZ::Ω, ΔdivJ, divKcs, ΔpleC, ΔcpaE or ΔpodJ cells. Expression of Dendra2-ZitP or Dendra2-CpaM was induced from the chromosomal xylX locus with xylose 4 hours before imaging. Scale bars: 1 µm. (G) (H) Epifluorescence (Dendra2) and Nomarski (DIC) images depicting the localization of the motility-deficient and pilus-proficient Dendra2-ZitPCS variant (G) or the motility-proficient and pilus-deficient Dendra2-ZitP1-133 variant (H) in ΔzitP cells. Arrow heads pinpoint stalked poles. We induced expression of Dendra2-fusions from the xylose-inducible Pxyl promoter on a plasmid integrated at the chromosomal xylX locus by the addition of xylose 4 hours before imaging. Scale bars: 1 µm.
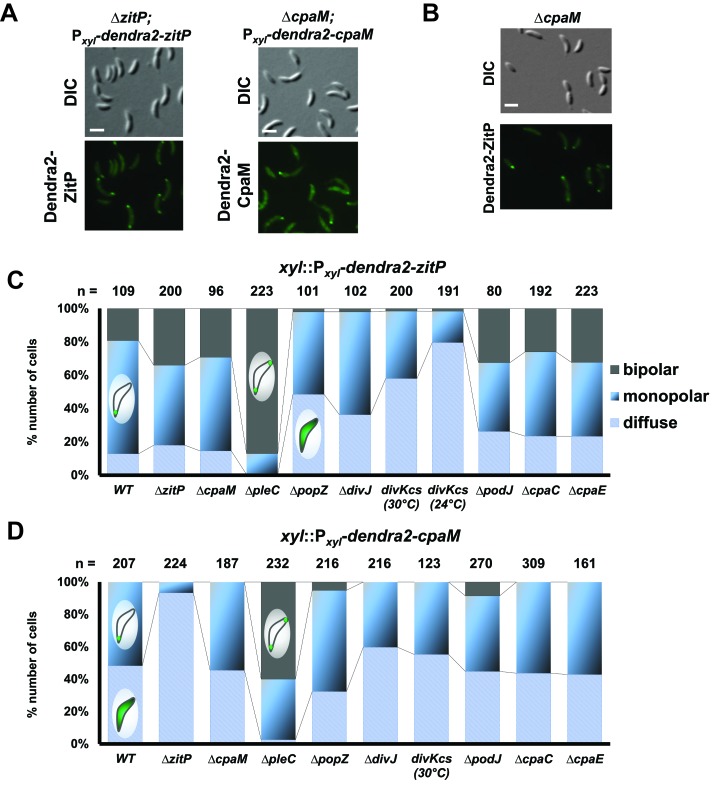
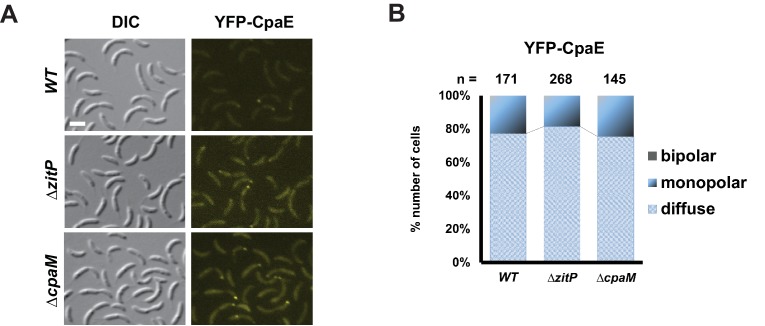
Figure 3—figure supplement 1. Extrinsic determinant for the localization of ZitP and CpaM.
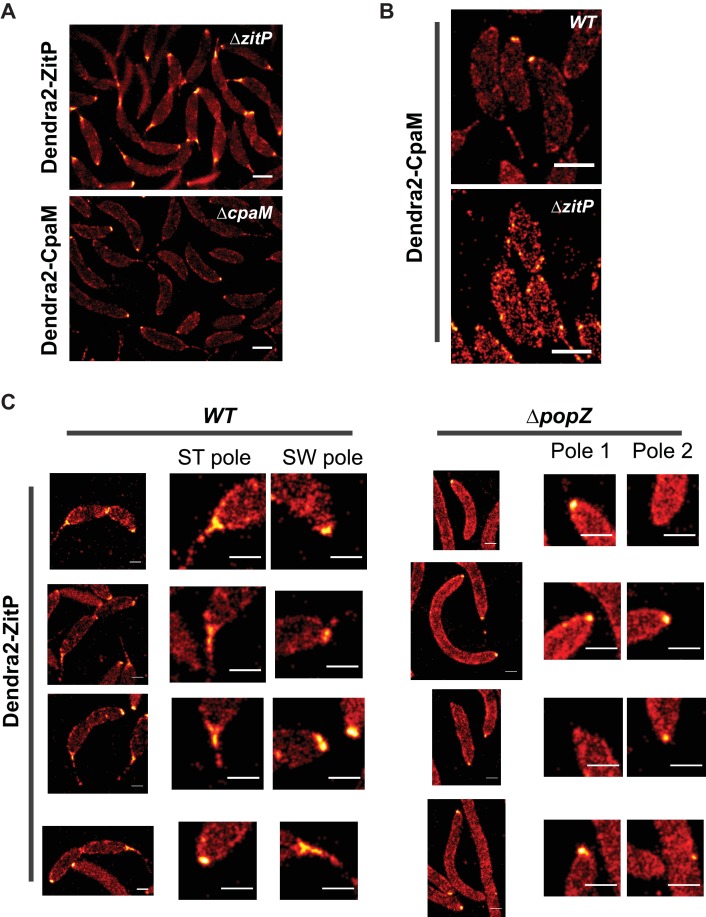
Figure 3—figure supplement 2. ZitP and CpaM polar localization by PALM.
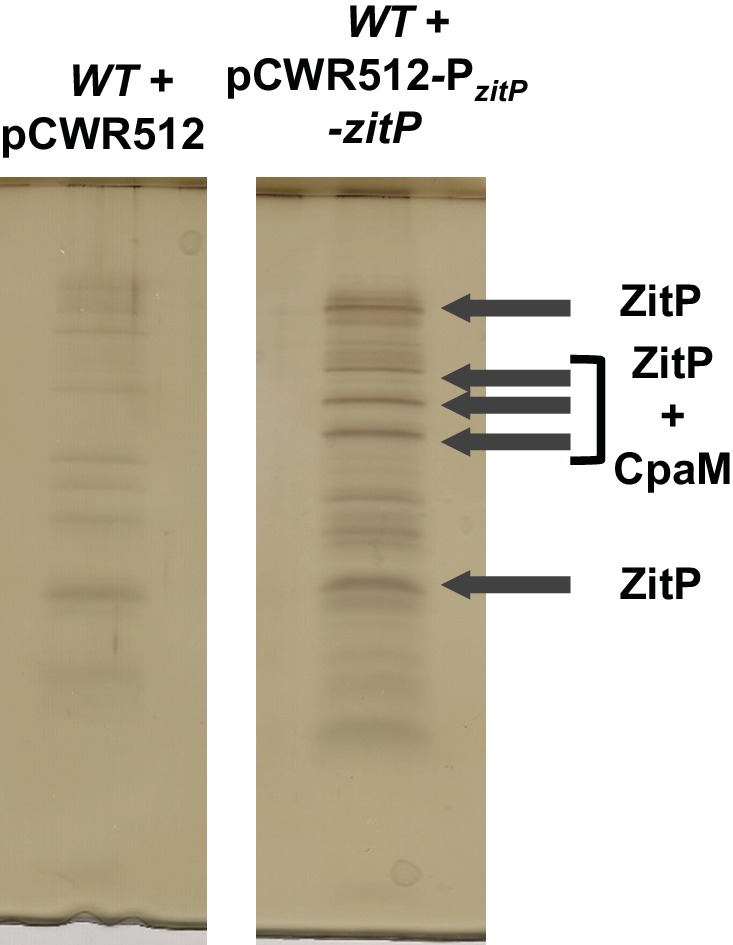
Figure 3—figure supplement 3. Tandem affinity purification of ZitP.