Fig. 1. Systematic mapping of genetic variation that affects the gene-regulatory cascade.
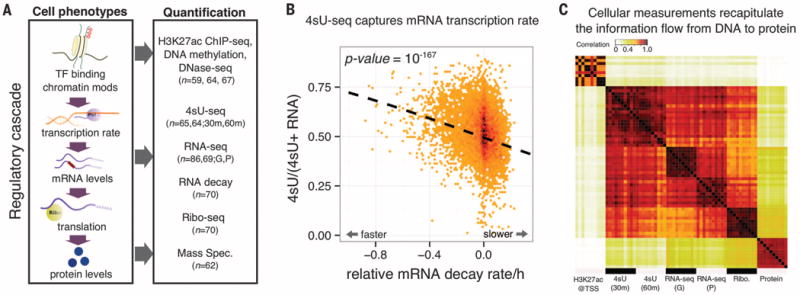
(A) QTLs mapped for eight cellular phenotypes in LCLs. For 4sU, 30 m and 60 m refer to different measurement time points. For RNA-seq, G and P refer to data from two studies (6, 17). TF, transcription factor. (B) Steady-state RNA levels reflect a balance between transcription and decay. Normalized mRNA decay rates (x axis) are plotted against the ratio of new mRNA to steady-state mRNA (y axis). Each data point is a gene. (C) A correlation matrix of seven data sets reflects the expected order of steps in gene regulation. Each entry in the matrix shows the correlation across genes between measurements of a pair of samples and/or data types. The plot shows 10 different random samples for each data type.
