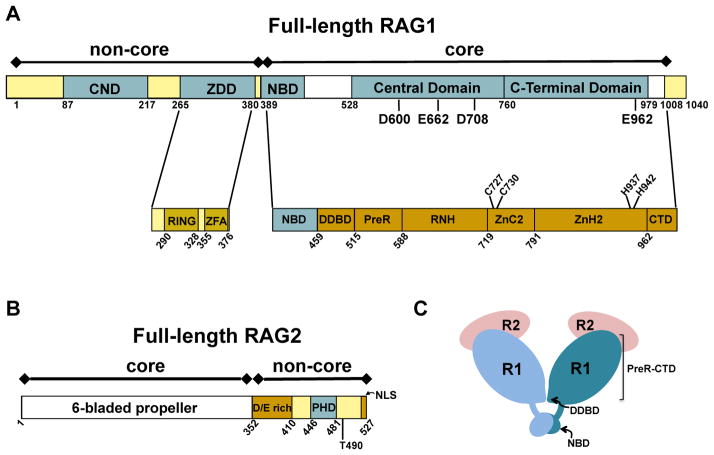
Figure 2. Schematics of domains and subdomains in the RAG proteins.
Core and non-core regions are denoted above the bar diagram of each RAG protein. (A) Full length murine RAG1. Non-core regions in RAG1 include an N-terminal region (residues 1-388) and a short C-terminal region (residues 1009-1040). Topologically-independent domains are shown as blue boxes. In non-core RAG1 these include the central non-core domain (CND) and zinc dimerization domain (ZDD). A crystal structure of the ZDD showed two sub-domains, the RING finger and zinc finger A (ZFA). Domains in core RAG1 include the nonamer binding domain (NBD), the Central domain, and the C-terminal domain. The core region contains multiple modules, as labeled in the lower bar. The three DDE active site residues (D600, D708, and E962), a putative fourth active site residue (E662), and zinc-coordinating ligands (C727, C730, H937, and H942) are labeled in the RAG1 bar diagrams. (B) Full length murine RAG2. The core region is a 6-bladed propeller domain. Non-core RAG2 contains an acidic region, labeled as D/E rich, as well as a plant homology domain (PHD) that specifically binds the trimethylated lysine residue of histone H3K4me3. Positions of residue T490 and the C-terminal nuclear localization signal (NLS) are indicated. (C) Cartoon depicting the core RAG12RAG22 complex. RAG1 (R1) subunits are in light and dark blue, and RAG2 (R2) subunits are in pink. The domains (NBD and DDBD) that form the RAG1 dimer interface are labeled. The remaining subdomains (PreR to CTD) compose the catalytically active region of core RAG1.