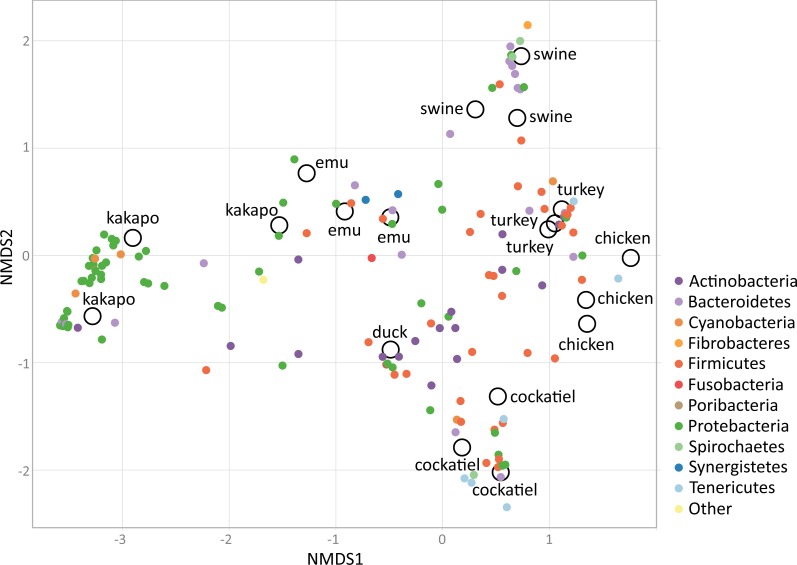
Figure 5. Non-metric dimensional scaling (NMDS), using Bray–Curtis dissimilarities, biplot analysis for the bird microbiomes.
There is no cluster attraction due to the sequencing technology used or 16S rRNA gene region sequenced. Swine microbiomes are used for outgroup purposes only, and they are clustered apart owing to their Bacteroidetes. Interestingly, the ordination has most Proteobacteria on the left, and Firmicutes on the right quadrant. There are clusters of poultry-related samples of different species, such as the case for turkey, chicken, and emu. Kakapo are clustered separately from every other species on the left quadrant and are clearly being clustered by its Proteobacteria abundance. The wild duck also clusters separately owing to its particular microbiome configuration with Actinobacteria, Fusobacteria, and Proteobacteria. Finally, cockatiels are in the bottom right quadrant, being clustered separately owing to their Firmicutes, Tenericutes, Spirochaetes, and some Proteobacteria families. The stress level for NMDS = 0.1429879. This is a coarse comparison at phyla level between microbiome samples of different species, fine detail could be obtained only through comparing samples using the same sequencing technology, and coverage.