Abstract
Current understanding of environmental cross-talk with genetic makeup is found to be mediated through an epigenetic interface which is associated with prominent reversible and heritable changes at gene expression level. Recent emergence of epigenetic modulation in shaping the genetic information has become a key regulatory factor in answering the underlying complexities associated with several mental disorders. A comprehensive understanding of the pertinent changes in the epigenetic makeup of suicide phenotype exhibits a characteristic signature with the possibility of using it as a biomarker to help predict the risk factors associated with suicide. Within the scope of this current review, the most sought after epigenetic changes of DNA methylation and histone modification are thoroughly scrutinized to understand their close functional association with the broad spectrum of suicide phenotype.
Keywords: Suicide, epigenetics, DNA methylation, histone modification, gene expression
1. Introduction
Suicide is described as a person’s self-inflicted violence with a serious intent to obliterate life, which most of the time culminates into serious physical injuries (CDC, 2015). Under the current socio-economic scenario, every 40 seconds a person dies by suicide somewhere in the world and many more attempt to commit suicide (WHO, 2014). Notably, in the young age group between 15–29 years, suicide is the second leading cause of death globally, whereas, it is attributed as the tenth leading cause of death across all ages (CDC, 2015). These facts and the lack of timely implementation of proper preventive strategies make suicide a global health problem that warrants a serious global imperative to tackle it (WHO, 2014).
Present understanding of suicide and its underlying etiology has pointed out the involvement of several risk factors collectively emphasizing the role of genetic makeup, history of early life events, and the current psychopathological status of the victim (Turecki, 2014). With the exception of the inheritability of the genetic constitution, the other two state-dependent factors (i.e. early life events and recent psychopathological status) are the underlying causes of suicidal behavior (SB) found to be mostly vulnerable to environmental stimuli (Mann and Haghighi, 2010).
Over the last decade, much work has been done to understand the genetic basis of suicidal behavior. While collecting support for the genetic basis of suicidal behavior, a number of studies suggest the familial aggregation of suicide, which indicate that the likeliness of attempting or committing suicide is 10 times higher in relatives of suicide completers (Brent and Melhem, 2008; Kim et al., 2005; McGirr et al., 2009). Regardless of psychiatric disorders, this risk remains high within families with a history of suicidal attempts or incidences (Kim et al., 2005). Additional twin and adoption studies find that the predisposition to suicidal behavior is also partly heritable (Roy and Segal, 2001; Roy et al., 1991; Roy et al., 1995). Although estimates of heritability for suicide among twins range from 21% to 50% (Currier and Mann, 2008), general population-based studies extend this limit up to 55% considering the broader aspects of suicidal behavior. Moreover, similar influence of environmental factors has been reported in twins where suicide concordance rate is 5.6% (1/18) in monozygotic (MZ) twin versus 0% (0/21) in dizygotic (DZ) twins (Kallman, 1953). Another study shows a similar pattern with an increasing incidence of suicide in 20% MZ and 3% in DZ (Pedersen and Fiske, 2010). Therefore, the involvement of genetic components is more likely to increase the risk of suicide incidences with a strong background of heritable predisposition. Likewise, a notable magnification of suicidal incidences was documented in the biological relatives of adoptees who die by suicide compared with adoptive relatives (Slap et al., 2001). Although a considerable amount of interest has been drawn to understand genetic predisposition as a major contributing factor in suicidal behavior, the neurobiological factors associated with suicide remain inconclusive.
Advancement in genome-wide expression studies has paved the way to unfold the potential involvement of a list of candidate genes and their improper functioning as a probable outcome or as a risk factor for suicidal behavior (Mann et al., 2009). Involvement of serotonergic, dopaminergic, and to some extent, noradrenergic systems have been investigated to explore their genetic association with the complexity of suicide (Zai, 2012). However, the involvement of multiple genetic factors and the heterogeneity of interacting genetic loci makes it more complicated to interpret the complex phenotype of suicidal behavior and has even failed to indicate a strong association from genome-wide association study (Galfalvy et al., 2015).
In this context, few recent findings have shown promising results in interpreting the missing link between the heritability of suicidal behavior and the interaction between environment and genome (Mandelli and Serretti, 2016; Roy, 2012) from a completely new perspective of epigenetic modulation (Bani-Fatemi et al., 2015; Lockwood et al., 2015). This type of mechanistic regulation and switching does not involve any permanent change in DNA sequence and seems to be transient in effect (Labonte and Turecki, 2010) as well as consistent with the episodic modulation observed in suicidality and suicidal ideation. Thus, epigenetic modifications including histone modification and small non-coding RNA-mediated post-transcriptional gene regulation in addition to DNA methylation are capable of causing a fast and quick response at the cellular level with the corresponding changes in environmental cues (Kubota et al., 2012).
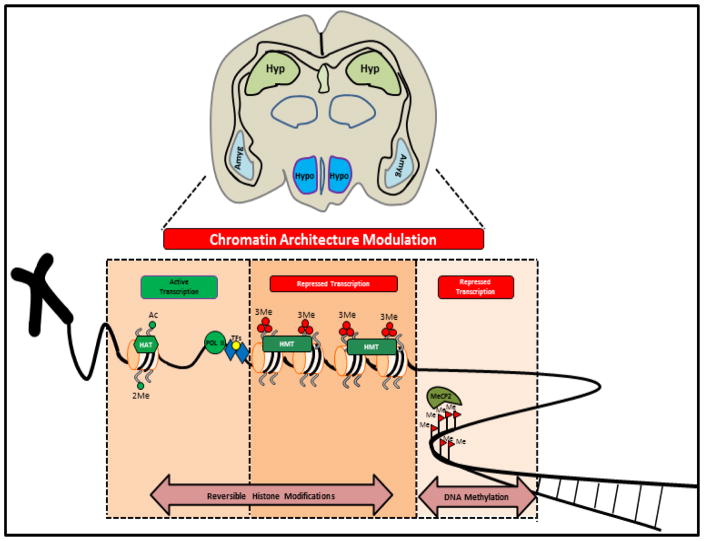
Suicidal behavior is primarily dictated by the predisposed genetic constitution of the individual and most likely to be orchestrated by the epigenetic milieu working in continuum with environmental inputs. It has consistently been shown that suicidal behavior is mostly affected by psychiatric disorders (Harris and Barraclough, 1997) where epigenetic perturbation of underlying neuro-molecular circuitry principally involves altered DNA methylation and histone modifications (Figure 1) (Turecki, 2014).
Figure 1.
Schematic representation showing possible epigenetic regulatory mechanisms proved to impart dynamic alteration in chromatin architecture primarily responsible for reversible switching of transcriptional status within the genomic context of suicidal brain. Covalent attachment of acetyl group or di-methylation on H3K4 residue marks the genome for active transcription with recruitment of RNA PolIII enzyme and associated transcription factors (TFs). Selective tagging of histone tails with tri-methyl groups (e.g. H3K9) mostly restricts the genomic milieu from active transcription. Similar situation prevails when reversible covalent modification of cytosine residues with methyl group transforms the chromatin organization to become repressive. These two fundamental repressive mechanisms (histone modifications and 5′ cytosine methylation) are mostly found to be dynamic in silencing candidate genes in specific brain regions of suicide completers or peripheral tissues like blood mono-nuclear cells of suicide attempters or ideators.
In this review, we have attempted to critically evaluate the present understanding of epigenetic regulation in determining the phenotypic outcome (Table 1) and severity of suicidal behavior (McGowan and Szyf, 2010); we have an extended objective to explore the future possibility of its usefulness as suicide biomarker with strong diagnostic value and target for effective therapeutic strategy.
Table 1.
Snapshot of Suicide Epigenetics
| Tissue Type | Type of Epigenetic Modifications | Outcome | Genes Involved | Case (N) | Control (N) | Publication |
|---|---|---|---|---|---|---|
| Post-mortem Brain | DNA methylation in frontopolar cortex | Promoter hypermethylation | GABAA α1 | 10 | 10 | Poulter et al., 2008 |
| DNA methylation in frontal cortex (BA 8/9) | Promoter hypermethylation | TRKB.T1 | 10 | 10 | Ernst et al., 2009 | |
| DNA methylation in hippocampus | Promoter hypermethylation | NR3C1 Exon 1F | 24 (12 = SNA, 12 = SA) | 12 | McGowan et al., 2009 | |
| DNA methylation in orbitofrontal cortex (BA11) | Unaltered promoter methylation | QKI Exon 1 | 8 | 8 | Klempan et al., 2009 | |
| DNA methylation in prefrontal cortex (BA 8/9) | Unaltered promoter methylation | SMS Exon 1 and SMOX | 10 | 10 | Fiori and Turecki, 2010 | |
| DNA methylation in Wernicke area | Promoter hypermethylation | BDNF promoter Exon 4 | 44 | 33 | Keller et al., 2010 | |
| DNA methylation in prefrontal cortex (BA 8/9) | Promoter hypermethylation at polymorphic site | SAT1 | 10 | 10 | Fiori and Turecki, 2011 | |
| DNA methylation in Wernicke area | Unaltered promoter methylation | TRKB.T1 | 18 | 18 | Keller et al., 2011 | |
| DNA methylation in hippocampus | Promoter hypermethylation | NR3C1 Exon 1C and 1H | 42 (21= SNA, 21= SA) | 14 | Labonte et al., 2012 | |
| DNA methylation in BA 44 | Promoter hypermethylation in ARG2 and AMD1 | OAZ1, OAZ2, AMD1, and ARG2 | 33 | 31 | Gross et al., 2013 | |
| DNA methylation in hippocampus (Dentate Gyrus) | Genome wide altered methylation | NR2E1, CHRNB2, GRM7 and DBH | 46 | 16 | Lebonte et al., 2013 | |
| DNA methylation in prefrontal cortex (BA 8/9) | 3′UTR hypermethylation | TRKB.T1 | 11 | 13 | Maussion et al., 2014 | |
| DNA methylation in prefrontal cortex | Promoter, 3′UTR hypermethylation | SKA2 | 3 independent cohorts | Guintivano et al., 2014 | ||
| DNA methylation in prefrontal cortex (BA 8/9) | Genome wide (Intragenic DMR hypermethylation) | GRIK2, BEGAIN | 22 | 17 | Nagy et al., 2015 | |
| DNA methylation in hippocampus | Genome wide | ALS2, DGKZ, HCK | 25 | 16 | Labonte et al., 2012 | |
| DNA methylation in prefrontal cortex | Genome wide | EYA2, MEGF11, LMNA, GLUD1, ERBB3 | 25 | 28 | Haghigi et al., 2014 | |
| Histone modification in orbito frontal cortex (BA 10) | Promoter based increased H3K27 trimethylation | TRKB.T1 | 20 | 10 | Ernst et al., 2009 | |
| Histone modification in prefrontal cortex (BA 8/9) | Unaltered promoter based H3K27 trimethylation | SMS and SMOX | 10 | 10 | Fiori and Turecki, 2010 | |
| Histone modification in prefrontal cortex (BA 8/9) | Unaltered promoter based H3K27 trimethylation | SAT1 | 10 | 10 | Fiori and Turecki, 2011 | |
| Histone modification in prefrontal cortex (BA4/17) | Promoter based increased H3K9me3 | CX30, CX40 | 22 | 22 | Nagy et al., 2016 | |
| Histone modification in prefrontal cortex (BA 44) | Promoter based increased H3K4 trimethylation in OAZ1 | AMD1, ARG2, OAZ1, and OAZ2 | 34 | 34 | Fiori et al., 2012 | |
| PBMC | DNA methylation in leukocytes | Promoter hypermethylation | BDNF | 108 | – | Kang et al., 2013 |
| DNA methylation in leukocytes | 3′UTR hypermethylation | SKA2 | 3 independent cohorts | Guintivano et al., 2014 | ||
| DNA methylation in leukocytes | 3′UTR hypermethylation | SKA2 | 466 total subjects | Sadeh et al., 2016 | ||
| DNA methylation in leukocytes | Exon Oriented hypermethylation | HTR2A | 32 | 35 | Bani-Fatemi et al., 2016 | |
| DNA methylation in leukocytes | Promoter hypermethylation | BDNF | 732 | – | Kim et al. 2014 | |
BA = Brodmann Area; SNA = Non Abused Suicides; SA = Abused Suicide; UTR = Untranslated Region; H = Histone; K = Lysine; PBMC = Peripheral Blood Mononuclear Cell.
2. Influence of DNA methylation based (5mC) epigenetic modifications in suicide pathophysiology: knowledge from post-mortem brain studies
DNA methylation is the most studied epigenetic modification in the mammalian genome where covalent attachment of methyl group to cytosine residues by DNA methyltransferase enzyme renders a substantial change in information processing during gene expression (Moore et al., 2013). This particular type of DNA-based modification is classically found to be more stable and commonly present on adjacent CpG dinucleotide in the mammalian genome. Though much work has been done to understand the function of CpG methylation in relation to gene regulation which are generally found to be repressive in nature, it is inconclusive to suggest a negative correlation between the degree of CpG methylation and gene expression which holds partially true only when they are localized to promoter regions instead of gene body (Jones, 2012).
2a. Affecting neurons
An initial understanding of the monoaminergic system and their involvement in major psychiatric disorders has drawn attention for a long time; however, other possible molecular pathways appear to be critically and equally involved in the neurobiology of mental disorders (Fiori et al., 2011; Fiori and Turecki, 2012). Over the last few decades, extensive studies to elucidate the intricacies of dysregulated neurotransmission in psychiatric illnesses, potentially leading to suicidal behavior, have shifted the focus from monoaminergic hypothesis to other molecular pathways including noradrenergic, glutamatergic and γ-aminobutyric acid (GABA) neurotransmission, as well as the polyamine system in post-mortem brains (Fiori and Turecki, 2010).
A. Involvement of GABAergic system
One of the very first reports that elaborated on the role of DNA methylation in reprogramming the gene expression of suicide brain focused on the GABAergic system (Poulter et al., 2008). This report connects the brain region specific (frontopolar cortex) hyperactivity of DNA methyltransferase (DNMT-3B) gene with an observed hypermethylation status of GABAA α1 subunit in suicide brains. The inverse relationship shown in this study between specific DNA methyltransferase enzymes with site-specific methylation of GABAA α1 subunit was partly answerable as a possible underlying epigenetic regulatory mechanism to the earlier observation of GABAA α1 subunit downregulation in suicide brain (Merali et al., 2004). The physiological relevance of this GABAergic expression deficit could be attributed in assessing hedonic and decision-making capabilities of prefrontal cortex which was mostly found to be underserved in suicidal behavior as part of the impaired cognitive processes (Pabba and Sibille, 2016). But this report was rather inconclusive to establish a causal relationship between DNMT-3B expression, methylation, and the reduced mRNA abundance of the GABAA α1 subunit in suicide brain.
B. Involvement of polyamine system
Recent advancement in gene expression analysis with high-throughput technologies provided considerable evidence to closely examine the other systems and their extensive participation in disorders like major depression, schizophrenia and bipolar disorder which are potentially associated with suicidal phenotype (Hoertel et al., 2015). The identification of polyamine system, previously known for its involvement in the cellular cycle and cancer biology (Gerner and Meyskens, 2004), has gradually gained acceptance in the recent past because of its genetic and epigenetic association with suicide neurobiology (Fiori and Turecki, 2008).The very first identification of a downregulated enzyme ‘SAT1’ from this polyamine system across three major Brodmann Areas (BA 4, 8/9 and 11) in French-Canadian suicide completers added a substantial informative value to this less studied metabolic pathway for further investigation (Fiori et al., 2011). Owing to the fact of observing differential expression for SAT1 gene in selective brain regions of suicidal completers has warranted the need to closely examine the underlying regulatory mechanisms for this SAT1 gene and other genes actively participating in this pathway. Careful observations unveiled few more genes namely SMS, SMOX and AMD1 as differentially expressed candidates in suicide completers (Guipponi et al., 2009; Sequeira et al., 2006; Sequeira et al., 2007). With a growing need to underpin the functional relationship, Fiori and group (2010) hypothesized the possible subduing impact of promoter DNA methylation as a causal factor on observed attenuated expression of two of the previously mentioned polyaminergic genes, SMS and SMOX in prefrontal cortex of suicide completers (Fiori and Turecki, 2010). Brodmann area 8/9 based analysis of SMS and SMOX genes within 1kb upstream of their promoter region revealed either low level of methylation or complete unmethylated state in 10 cases of suicide completers. Furthermore, an overall methylation status analysis unfolds no significant difference between suicide completers and healthy control followed by the identification of a non-significant correlation between gene expression downregulation and promoter DNA methylation. However, a significant relationship was observed between SMOX expression and a specific site based hypermethylation on its promoter. The outcome of this study appeared to be rather surprising from DNA methylation standpoint, but it could possibly be explained considering the very small population size which might have less ability to resolve the distinctive effect of methylation alteration (Fiori and Turecki, 2010).
Further substantiation for the involvement of polyaminergic system in suicide neurobiology and their epigenetic perturbation by DNA methylation came from an elegant study conducted on suicide completers (Fiori and Turecki, 2011). In this study, importance was given to the gene SAT1, synthesizing spermidine/spermine N1-acetyltransferase, a rate-limiting enzyme in polyamine catabolism from polyamine system in the brain. A serious attempt was made to understand the role of other gene regulatory mechanisms instrumenting the downregulation of SAT1 gene in suicide completers, except two regulatory single nucleotide polymorphisms in its promoter. The study provided further insight into the functional impact of promoter methylation on SAT1 gene expression in prefrontal cortex by mapping the presence of a polyamine responsive element (PRE) within the two heavily methylated sites. The PRE is primarily involved in increasing the SAT1 expression by interacting with transcription factor Nrf2 in conjunction with polyamine-modulated factor-1 (PMF-1). Additionally, the study turned out to be more interesting in characterizing an SNP (rs6526342) within the SAT1 gene promoter with high methylation incidence in suicide completers. A frequent occurrence of the C allele at this rs6526342 locus in suicide completers potentially draws the possibility of site-specific methylation induced SAT1 repression in causing suicide pathophysiology (Fiori and Turecki, 2011).
More recent identification of additional polyamine pathways related genes including ornithine decarboxylase antizymes 1 and 2 (OAZ1 and OAZ2), arginase 2 (ARG2), and AMD1and their significantly induced transcriptional output in BA 44 of suicide completers with mood disorder background (Gross et al., 2013) has reinforced the involvement of differential regulatory mechanism on these rate limiting enzyme syntheses. In fact, identification of a hypomethylated status in the promoter area of AMD1 and ARG2 correlated well with the observation of upregulated expression of these two genes in suicide completers. In spite of observing an increase in ORG1 and 2 gene expression and significant alteration in their promoter methylation, the study failed to show any significant correlation between these two observations. This observed inadequacy could partly be explained because of the smaller (500 bp) promoter fragments investigated for methylation study and might have been minimally involved in OAZ1 and 2 genes expression regulation. Nonetheless, this study provides a strong convergent evidence for the role of DNA methylation as a potential epigenetic modulator in orchestrating the various genetic components of polyamine system as evidenced by other reports collectively influencing the suicidal endophenotype (Fiori and Turecki, 2010; Limon et al., 2016). Furthermore, this study (Gross et al., 2013) conveys additional information on DNA methylation based epigenetic marks that could possibly be considered an exclusive signature of suicidal behavior. As part of this study, the identification of significantly different epigenetic marks on gene promoters in suicide completers could be explained as exclusive methylation signature capable of rendering pathophysiological consequences. The underlying uniqueness of this suicide-specific epigenetic study is the inclusion of suicide subjects with various mental disorders. Out of 34 subjects, 28 were diagnosed with axis I disorders and comprised of heterogeneous population of 3 schizophrenic cases, 12 major depressive disorder (MDD) cases, 7 bipolar cases and 3 substance abuse cases (Gross et al., 2013). Despite this heterogeneity, a significant observation of hypomethylated status of ARG2 and AMD1gene promoters with negative correlation to their gene expression makes it compelling to associate them with suicidal behavior. Similar associations have been found by Keller et al. (2010) where a consistent observation of DNA hypermethylation BDNF has been linked to suicide phenotype while analyzing the cortical brain area from 44 suicide completers with a spectrum of psychiatric diagnosis. This study is discussed in more detail in the next section: “Involvement of neurotrophic system” (Keller et al., 2010).
C. Involvement of neurotrophic system
A paradigm shift in normal mental states involving cognition, emotion, and memory have been considered as cardinal signs to acknowledge the representation of suicidal behavior (van Heeringen, 2003) with a close relationship to the pathophysiology of mood disorder characteristically documented with the dysfunctional serotonergic system with a hippocampal origin (Mann, 2013). However, a close examination of suicide prefrontal cortex with special reference to area BA46, mostly involved in affective disorders of altered mood, has been shown to be overrepresented in suicide behavior (Hoptman et al., 2002; Kamali et al., 2001; Keilp et al., 2001; Mann, 2003; Raine et al., 1998; Raust et al., 2007). The brain’s inability to respond properly to environmental stimuli due to impaired structural and neural plasticity has been well-established to cause suicide pathogenicity (Dwivedi, 2009, 2010; Furczyk et al., 2013). Recent neuroimaging identification of orbitofrontal and dorsolateral prefrontal cortex based structural abnormalities in suicide victims further reinforces the importance of impaired neuronal plasticity (Ding et al., 2015; van Heeringen et al., 2011). An overwhelming concern and ever-increasing need to better apprehend this compromised structural and functional plasticity, eventually unfolded the role of brain-derived neurotrophic factor (BDNF) and its cognate receptor molecule tropomycin receptor kinase B (TRKB) in suicide neurobiology (Dwivedi, 2009, 2010; Dwivedi et al., 2003; Lee and Kim, 2010; Pandey et al., 2008). An in-depth study deciphering the functional contribution of BDNF gene in neuronal growth, differentiation, and maintenance during developing as well as in adult brain undoubtedly acknowledged its pivotal role in shaping the structural, morphological, and synaptic plasticity (Cohen-Cory et al., 2010; Park and Poo, 2013). Therefore, it is not difficult to envisage that alteration in the functionality of BDNF gene due to abnormal gene expression or impaired mature protein processing and transportation may give rise to serious pathological conditions abrogating the entire cellular pathway related to plasticity (Autry and Monteggia, 2012; Lipsky and Marini, 2007; Martinowich and Lu, 2008; Martinowich et al., 2007). Similarly, an equal contribution was noticed from TRKB receptor where its abstinence from proper functioning may turn into abnormal BDNF-TRKB intra-cellular signal transduction function with a loss of neuroprotective effect on neurons and glial cells (Minichiello, 2009; Rose et al., 2003). Converging lines of evidence further indicated significantly diminished expression of both BDNF and TRKB genes in prefrontal cortical neurons as well as hippocampus of adult suicide subjects where neuronal plasticity has been shown to be heavily compromised (Ernst et al., 2009; Karege et al., 2005).
The importance of area based functional compartmentalization in brain regions for epigenetic study became evident from a report by Keller et al. (2011) (Keller et al., 2011). This group showed an interesting result on TRKB and TRKB.T1 (truncated form of TRKB) expression and promoter methylation in Wernicke’s area which did not correlate with suicidal behavior unlike the previous report for the same gene in different Brodmann area (BA 8/9) (Ernst et al., 2009). TRKB receptors are found to be expressed in three different isoforms: the splice variant TRKBFL codes for a full-length receptor with functional tyrosine kinase domain (TKD), whereas the other two truncated isoforms TRKB.T1 and T2 are devoid of any functional TKD and do not participate in triggering a fast intracellular signaling response unlike their full-length counterpart (TRKB FL). The truncated prototype TRKB.T1 is predominantly expressed in brain astrocytic population with a defined role in calcium release from intracellular storage after receiving a brief exposure to BDNF (Rose et al., 2003). However, a subtle difference lies between the TRKBFL and TRKB.T1 receptors in utilizing two different channels to evoke the intracellular calcium response once exposed to BDNF-ligand (Carim-Todd et al., 2009). Interestingly, preclinical studies in TRKB.T1 deficient rodent model have indicated its role in developing anxiety-like behavior in comparison with normal littermates (Carim-Todd et al., 2009). Such compelling evidence may be contradictory to the unlikely outcome of the study conducted by Keller and colleagues and possibly indicating a bar due to smaller population size (n = 18/group). Although the study was based on a small sample size and limited to Caucasians, this finding was interesting to understand the concept of brain area specific epigenetic signature in conferring characteristic gene expression pattern relevant to suicidal behavior. Furthermore, a detailed examination in same samples, focused again on Wernicke’s area, deciphered a significant hypermethylation status of BDNF Exon IV promoter in suicide completers. This finding appeared to be in agreement with the previous report from the same group (Keller et al., 2010) where a subset of the same samples were used to study the BDNF exon IV methylation pattern. BDNF promoter methylation in Wernicke’s area from human brain was a significant step to better understand the underlying epigenetic complexity of suicide. Wernicke’s area of prefrontal cortex, grossly representing BA 21 and 22, is better known for semantic thinking and decision making which is mostly affected in suicidal behavior (Jollant et al., 2005; Jollant et al., 2007; Jollant et al., 2010; Keller et al., 2010). Identification of BDNF exon IV specific hypermethylation status in Wernicke’s area invaluably highlights the serious outcome of its deficit as neurotrophic factor and possibly correlates the deficiency with aforementioned functional impairments in associative and neurocognitive processing of suicidal brain (Jollant et al., 2005; Jollant et al., 2007; Speckens and Hawton, 2005).
In 2014, Maussion et al. provided a deeper insight into understanding the methylation-based TRKB.T1 gene regulation in suicide brain with a completely different perspective (Maussion et al., 2014). Considering 3′UTR coding sequence for this gene as the potential site for DNA based methylation, the study was able to correlate astrocyte restricted TRKB.T1 isoform expression downregulation with suicide pathophysiology. The study was mainly focused on exploring the possibility of relative methylation contribution in regulating astrocyte-based TRKB.T1 isoform expression in frontal cortex of suicide completers emphasizing the involvement of four CpG sites in 3′UTR coding part of TRKB gene. This regulatory mechanism was different from miRNA-185* mediated post-transcriptional repression of TRKB.T1earlier found to happen in the frontal cortex of suicide brain (Maussion et al., 2012). Unlike the promoter oriented methylation regulation on TRKB.T1 isoform (Ernst et al., 2009), this study added a new dimension by considering 3′UTR coding DNA as the target of methylation modification to answer the high magnitude of transcriptional repression observed for TRKB.T1 isoform in frontal cortical areas of suicide completers. The identification of additional polymorphic CpG site located in close proximity to other four sites failed to produce a significant association with suicide after genotyping 33 suicide completers. This observation of site-specific differential methylation pattern with a negative effect on corresponding transcript expression was supported by the mapping of CTCF binding site overlapped with four CpG sites, which is in agreement with the observed distribution of 1.42% 3′UTR specific CTCF binding across the whole eukaryotic genome apart from 5.34% promoter specific distribution (Pena-Hernandez et al., 2015). Most likely, the characterization of a CTCF binding site in the same area of preoccupying methylated CpG sites at the 3′UTR transcriptional unit of TRKB.T1 holds a strong possibility towards abated interaction of CTCF within this intragenic region. Moreover, the study is also in agreement with other previous findings where methylation in the 3′UTR coding parts other than the promoter region has been shown to be involved in transcript specific regulation of gene under the tight spatio-temporal cellular environment (Choi et al., 2009; Zheleznyakova et al., 2015). While signifying the relevance of astrocyte-specific TRKB.T1 isoform expression modulation in suicide completers, the above mentioned mechanistic intricacies hold true to justify the observed epigenetic interference possibly linking the astrocyte-mediated neuronal activity modulation (Allen, 2014) found to be affected in suicide brain possibly via TRKB.T1 Ca2+ signaling (Rose et al., 2003).
D. Involvement of hypothalamus-pituitary-adrenal (HPA) axis
Consistent reports on blunted HPA axis functionality has elaborated the importance of stress originated maladaptive neuroendocrine system as one of the major risk factors behind suicidal behavior, despite the involvement of other cellular and biochemical determinants such as neurotransmitters, neurotrophic factors and cytokines (Carballo et al., 2008; Melhem et al., 2016). Dysregulated glucocorticoid receptors (GR) are mostly found to be the cause of dampened negative feedback regulation mediated by glucocorticoid hormone in ascribing a hyperactive HPA axis response under stress environment (Arnett et al., 2016; Pandey, 2013). Complex molecular understanding of this maladaptive HPA axis response in adult suicide victims indicated an interesting link between their adverse early life experiences with dysregulated GR activity in hippocampi (Kundakovic and Champagne, 2015; Labonte et al., 2012). Similar kind of childhood adversity was found to be influential in regulating HPA axis functions in rodent model where deviation from normal maternal care in the form of low licking-grooming and arched back nursing on pups resulted in altered DNA methylation profile of GR promoter in hippocampus (Weaver et al., 2004; Weaver et al., 2007). This altered methylation at the earliest phase of life (first week after birth) worked as a stable epigenetic signature to heterochromatinize the conformation of GR promoter and impeded the binding of NGF1-A transcription factor in adulthood (Hellstrom et al., 2012). Thus, the involvement of diversified environmental insults like childhood abuse, maternal separation or nutritional deprivation received at earliest phases of life might be considered as risk factors to intensify suicidal behavior in adult phases of human life with altered stress responsiveness due to dysregulated HPA axis function. In 2009, a seminal study conducted by McGowan and colleagues contemplated the significant decline in hippocampal NR3C1 expression in 12 suicide victims with a history of childhood abuse (McGowan et al., 2009). Conversely, differences in glucocorticoid receptor expression was found to be absent in the same brain area of 12 suicide victims without a history of childhood abuse when compared with 12 controls. More surprisingly, this study was further comparable with the findings in rodent model as discussed earlier (Weaver et al., 2004; Weaver et al., 2007) showing similar changes in NR3C1 exon 1F promoter methylation closely associated with childhood adversity in suicide victims which most likely mirrors the incident of interrupted mother-infant interaction in rat model. Moreover, the observations made by Labonte and associates (Labonte et al., 2012) reiterated similar kind of site-specific DNA methylation changes in the vicinity of transcriptional start sites of three non-coding GR transcript variants (1B, IC and 1H) in hippocampai of suicide completers with a history of childhood abuse and found to be associated with decreased expression. Successive use of in vitro cell line system with luciferase reporter assay further interpreted the functional role of promoter DNA methylation in GR 1C transcript expression regulation; however, critical examination of anterior cingulate cortex from the same suicide subjects did not reveal any significant expression alterations of three GR transcript variants mentioned earlier (Labonte et al., 2012). This report represents the involvement of certain brain regions with compromised HPA axis functionality as risk factor for suicidal behavior. Altogether, these observations confirm the in vivo finding of coordinated DNA methylation response as stable epigenetic modification acquired on multiple promoters of the hGR gene during childhood adversities contributing toward decreased hGR expression and HPA dysregulation in adult suicide victims. However, a far-reaching effect of promoter DNA methylation in hippocampi of 25 suicide completers with a history of early life trauma showed a bidirectional pattern on a genome-wide scale (Labonte et al., 2012). The significant bidirectional changes in DNA methylation included both hyper- and hypomethylated status of gene promoters functionally enriched for genes associated with neural plasticity. In sum, this genome-wide methylation study put additional emphasis on understanding the acquired methylation changes due to traumatic incidences in early life that could be perpetuated in late life with an increasing risk of suicide in the individuals who were abused during childhood (Haghighi et al., 2014; Labonte et al., 2012).
Abnormal functional response of glucocorticoid receptors as part of dysregulated HPA axis is not only moderated by their impaired cellular transcription but also found to be affected by their compromised availability in nuclear compartment as transcription factor (Tatro et al., 2009). In this regard, a gene named SKA2 (spindle and kinetochore (KT)-associated protein 2). involved in anaphasic movement of mitotic chromosome (Hanisch et al., 2006), has been identified as a molecular escort to help translocate glucocorticoid receptor (GR) from cytosol to nucleus where GR acts as a transcription factor (Rice et al., 2008; Sadeh et al., 2016).). A study conducted by Guintivano and colleagues (Guintivano et al., 2014) showed DNA methylation-mediated epigenetic mode of gene regulation with a polymorphic nucleotide rs7208505 at cg13989295 locus of SKA2 gene in neuronal population of autopsy brain. Further analysis of this polymorphic locus present at 3′UTR of SKA2 gene unfolded a rare involvement of an exclusive DNA methylation signature with functional implications in altering GR responsiveness predominantly in prefrontal cortex (PFC) of suicide subjects (Guintivano et al., 2014). Moreover, observations from peripheral blood tissue of three living cohorts were found to be aligned with the brain findings replicating a similar functional impact of SKA2 epigenetic mark on its own expression in relation to suicidal ideation (Guintivano et al., 2014). Further molecular insight in explaining the diminished SKA2 expression in a cohort of PFC collected from National Institute of Child Health and Human Development Brain and Tissue Bank for Developmental Disorders at the University of Maryland included two additional regions of CpG sites located on promoter area and 1st exon of SKA2 gene. As an additive effect, 40% reduction in SKA2 gene expression was found to occur via CpG methylation of those two additional sites along with the epigenetic and genetic variation contributed by rs7208505 site. Another interesting observation which appeared to be noteworthy from this study was the close linkage between genetic and epigenetic variability in conferring a high risk factor for ethnicity based suicidal phenotypes. The above statement happened to be the most likely explanation for the observed low allelic frequency for “C” containing allele at rs7208505 site in African American race (18%) than other ethnicities, possibly rendering the site to be less methylated (Guintivano et al., 2014). Therefore, certain SKA2 variations suggest a lower risk factor for suicidal phenotypes based on allelic discrimination in moderating the propensity of methylation modification at defined genetic locus. However, the cellular and molecular mechanisms underlying the associated risk of suicide with SKA2 genetic variation at rs7208505 site indicated a connection with defective glucocorticoid signaling which could be increasingly driven by CpG methylation intensity in response to stress factors. This was found to be true while analyzing the stress and anxiety related matrices of three living cohorts, where relative involvement of stress appeared to be influential on SKA2 epigenetic variation in progressing the successive stages of suicidal behavior i.e. suicidal ideation and suicidal attempt. Therefore, this SNP based epigenetic alteration of SKA2 in influencing HPA axis functionality of suicide brain was much more complex in interpreting the bimodal regulation of SKA2 on GR activity, overlaid with additional epigenetic features as evidenced from SKA2 promoter methylation and impaired microRNA 301a expression (Smalheiser et al., 2012) from the same intronic location. However, the overall outcome of this study suggests a genetic variation at 3′UTR of SKA2 gene accompanied by a signature methylation mark may mediate risk for suicidal behaviors that progress from ideation to attempt to suicide via GR alteration (Guintivano et al., 2014).
Stressful life events including early life trauma has been well described to cause a myriad of psychiatric disabilities involving a major functional impairment in hypothalamic-pituitary-adrenal axis (HPA) axis (Faravelli et al., 2012a; Faravelli et al., 2012b). As detailed before, a non-responsive GR feedback mechanism serves as the epicenter to transduce the related neuroendocrine changes into a stress related phenotype (Herman et al., 2005; Jankord and Herman, 2008; Smith and Vale, 2006). Since, stress is highly associated with suicidal behavior; biochemical studies have unveiled the involvement of a few more operational molecular components from this stress axis system (Currier and Mann, 2008). FK506-binding protein 51 (FKBP5/FKBP51), alternately known as FKBP5, is one of the intracellular components that modulates stress response by moderating the GR sensitivity toward cortisol binding (Binder, 2009). Detailed biochemical analysis from in vitro experiments with extended support from in vivo observations have proved the inhibitory role of FKBP5 in impeding the nuclear translocation of GR (Guidotti et al., 2013; Tatro et al., 2009). By interacting with heat-shock protein 90 (Hsp90), P23 protein, and other co-chaperones of the steroid receptor complex, FKBP5 interferes with the interaction of GR with cytoskeletal motor protein dynein to delay their retrograde nuclear translocation (Zannas et al., 2016). As a result, a diminished GR-dependent transcriptional activity has been found to occur in nuclear environment as part of the dysfunctional stress response. This underlying cellular physiology is closely associated with a number of stress-related neuropsychiatric abnormalities including MDD, post-traumatic stress disorder (PTSD) and severe cognitive disbalance which generally lead to suicidal phenotype (Perez-Ortiz et al., 2013; Zannas and Binder, 2014). Mounting reports have been published in past years where FKBP5 was made accountable for the debilitating condition either arising as suicidal thoughts or attempts in patients with a severe stress related background(Binder et al., 2008; Binder et al., 2004; Mehta et al., 2011; Roy et al., 2010; Roy et al., 2012; Willour et al., 2009). Identification of polymorphic loci on FKBP5 has recently been associated with suicidal incidences in European and Japanese population (Brent et al., 2010; Supriyanto et al., 2011). Childhood trauma-related suicidal attempts were found to be at higher risk due to the occurrence of specific transcript variant of FKBP5 gene in African American population (Klengel et al., 2013). Furthermore, FKBP5 variant-induced genetic predisposition was also evaluated as high risk factor for a wide spectrum of affective disorders ranging from bipolar disorder (BP), PTSD, to the recurring incidence of MDD (Appel et al., 2011; Menke et al., 2013; Willour et al., 2009; Zimmermann et al., 2011). However, a recent report on postmortem amygdala samples collected from 13 suicide completers with no clinical background of psychiatric disorders and free of psychoactive medication or substance abuse showed a significant depletion of FKBP5 gene expression both at transcript and protein level (Perez-Ortiz et al., 2013). This FKBP5 expression downregulation as well as reduction in GR gene product in amygdala of suicide victims could be a good source to interpret the experimental outcome which is free of any additive effects from comorbid psychiatric disorders and devoid of any artifacts generally contributed by medications prior to suicide. Most interestingly, the data from this study might be indicating the functional involvement of an ultrashort negative feedback loop that orchestrates the GR repression by FKBP5 and vice versa (Vermeer et al., 2003; Zannas et al., 2016). Other possibilities related to FKBP5 expression modulation, besides genetic factors, have been recently worked out in altered stress-gene interaction model where the involvement of epigenetic modulation via DNA methylation has been shown to be a regulatory factor (Ewald et al., 2014; Hohne et al., 2015). In this context, a report published in 2012 deserves to be mentioned briefly where the genetic predisposition contributed by a functional polymorphism in FKBP5 gene was shown to be fine-tuned by a DNA demethylation event to develop stress-related psychiatric disorder (Klengel et al., 2013). This study provided the clear mechanistic evidence of early life stress exposure in individuals carrying FKBP5 risk allele to receive an epigenetic re-patterning on glucocorticoid response element (GRE) of FKBP5, resulting in the disruption of ultra-short negative feedback loop between GR and FKBP5. This molecular disruption at intracellular level eventually got reflected in late life stress incidences functionally imbalancing the related hormonal system with the increasing risk for psychiatric disorders. Therefore, considering the relevance of neuropsychiatric disorders and their prevalence in suicide neurobiology, and given that FKBP5 plays a critical role in stress-related disorders, it will be interesting to examine if this gene is altered in suicide and if this alteration is due to epigenetic modifications like CpG methylation or histone modifications. However, earlier report on histone-based epigenetic regulation of FKBP5 has been documented in relation to cancer biology (Paakinaho et al., 2010), but its role is yet to be understood in suicide neurobiology.
Collective evidences from all these remarkable studies point out the significance of considering DNA methylation as interface of molecular crosstalk between predisposed genetic environment with external risk factors for assessment of complex suicidal behavior; however, they still fall short of interpreting the genome-wide involvement of DNA methylation alteration in impairing several biochemical pathways with increased vulnerability of interconnected neurobiological networks associated with suicide phenotype. Neuron-specific whole genome-wide assessment of refined DNA methylation signature in dentate gyrus of 46 male suicide completers from French Canadian descent interestingly unfolds a cohort of gene promoters significantly associated with cognitive process (Labonte et al., 2013). Functional clustering of such genes elaborates their role in learning, memory and other complex behavioral outcomes which characteristically denote the possible impairment of higher order function in suicide brains. Here in this study, more insightful analysis of four selected genes (NR2E1, GRM7, CHRNB2 and DBH) coding for membrane receptors and membrane-associated enzyme molecules with gross involvement in impulsivity, anxiety, learning and memory-related behavior exhibited a hypermethylation status concordant with their attenuated RNA level expression (except DBH). Overall deficit in the expression of these genes and their related functions in the hippocampus of suicide brains correlates with hyper-responsive behavioral phenotypes and are considered as predominant risk factors for increasing susceptibility to committing, suicide. In conclusion, this study proved to be an invaluable piece of first genome-wide differential DNA methylation report focused on neuronal subpopulation in hippocampal region of brains with substantial role found in dysregulating multiple cellular pathways intensifying the risk factors in predisposed suicide victims.
Taken together, all the evidences discussed herein undoubtedly point out the relative importance of post-mortem brain samples as effective functional model to study DNA methylation based epigenetic modification in understanding the suicide neurobiology with certain advantages. Most amenably, the post-mortem brain samples provide the expanded window for complete examination of the neurobiological changes at post suicidal phase (Furczyk et al., 2013; Pandey and Dwivedi, 2010). Moreover, because of the stable cytosine methylation, the differences in CpG methylation are unlikely to be a consequence of events immediately preceding death or during the postmortem period. Therefore, changes in brain pH do not affect DNA methylation (Ernst et al., 2008; Gross et al., 2013).
2b. Glial cell modulation
Identification of altered gene expression profile and their ontological relationship with astrocyte/oligodendrocyte apart from classically represented neurotransmitter function in fronto-cortical area has been a remarkable development to understand suicide neurobiology from a different perspective. These groundbreaking findings necessarily establish a strong cornerstone for central glial pathology in interpreting molecular complexities associated with depression and warrants further insight into their link with suicidal behavior (Fields, 2008; Klempan et al., 2009a; Sequeira et al., 2009).
Closer examination of affected gene list related to oligodendrocyte dysfunction in depressed suicide completers, regardless of the four cortical regions (BA 11, 44, 45 and 47) investigated by Klempan and colleagues (Klempan et al., 2009a), helped to sort out a gene named QKI with significant expression downregulation. QKI was studied to cause severe demyelination in CNS due to loss of function mutation and genetically found to be linked with schizophrenia showing a widespread expression deficit, which collectively points out its close functional relationship with mental disorders (Bockbrader and Feng, 2008). Moreover, to collect further substantial evidence in support of the documented expression deficit in QKI gene and the possible regulatory mechanism behind the compromised transcription, the same group (Klempan et al., 2009a) investigated the promoter methylation in cortical and subcortical brain regions of suicide completers with a history of depression. Despite finding a significantly repressed protein expression in BA 11 of suicide completers, the study was only able to identify traceable amount of CpG methylation in the same brain area. The week promoter methylation enrichment of QKI gene was resolved to be less than one per 80 CpG sites on a 281 base pair long genomic sequence investigated in BA 11 area of four suicide completers. The absence of any correlation between DNA methylation mediated promoter regulation on reduced expression of QKI gene both at mRNA and protein level might be suggesting the involvement of other epigenetic modifiers in those suicide completers. More likely explanation of histone modification impacting the transcriptional output and small non-coding RNA, like microRNA mediated translational impairment of QKI gene, could be connected with the oligodendroglial abnormalities observed in suicide brain which stems from neuropathology of major depression.
The role of astrocytes in evaluating suicide pathophysiology was further authenticated in a study by Nagy et al. in 2015. The group, for the first time, used a genome wide approach instead of candidate gene examination strategy to find out the degree of involvement of astrocyte specific differentially methylated regions (DMRs) in BA 10 area of patients who died by suicide (Nagy et al., 2015). The de-novo sequencing based identification of multiple DMRs across the genome from 76 suicide subjects helped to classify them based on their physical location regardless of promoter centric localization. Most interestingly, 90% of DMRs were found to be associated with non-promoter regions, and DMRs localized in the vicinity of gene body and intergenic regions exhibited most variability in methylation. Although significant functional relationship was demonstrated across the genome wide distributed DMRs with their variable DNA methylation status in suicidal brain, two DMRs were given thorough consideration because of their presence in the intragenic regions of GRIK2 and BEGAIN genes. GRIK2 was important in coding astrocyte associated glutamate ionotropic kainate receptor (Brand-Schieber et al., 2004). Improper functioning of this receptor to mediate calcium signaling under antidepressant treatment has been previously associated with depression (Hertz et al., 2014; Kawamoto et al., 2012). De-novo sequencing data from this study of Nagy and colleagues (Nagy et al., 2015), for the first time revealed a hypomethylation status of GRIK2 in intron 13 with a possible effect of producing alternate protein isoform with reduced permeability for calcium signaling in astrocytes. On the other hand, identifying BEGAIN gene with a hypomorphic functional status in astrocytes of suicidal brain more likely indicates a repressive effect exerted by the hypermethylated DMR characterized in its vicinity. Earlier report of a close physical association between BEGAIN protein and postsynaptic density protein (PSD95) (Yao et al., 2003) clearly marks its functional importance in post-synaptic transmission of astrocytes which appeared to be weakened under the glio-pathological background of suicide. The observation of dysfunctional astrocytes could be substantiated with the methylation alteration by DMRs, as mentioned before, to postulate a strong connection between astrocyte specific methylation signature with suicidal behavior.
The importance of site-specific methylation changes in the promoter region of genes and its strong correlation with suicide pathobiology was further exemplified in a study by Ernst and his colleagues (Ernst et al., 2009). The study correlated methylation specific changes in an astrocyte-specific transcript variant (T1) of TRKB gene with the MDD induced suicide behavior. A number of previous studies have shown a positive correlation between reduced TRKB expression with MDD pathology in the cortex (Qi et al., 2013; Tripp et al., 2012). However, the identification of two hypermethylated sites (site 2 and site 5) on TRKB promoter and the demonstration of a significant inverse correlation with the astrocyte-specific TRKB.T1 expression in suicide completers with an MDD background was first to be mentioned in this report (Ernst et al., 2009). Furthermore, this could be physiologically linked with impaired calcium signaling in the frontal cortex of suicide brain due to depleted synthesis of TRKB.T1 receptor, functionally known to elicit a neurotrophic response in cortical astrocytes, but the study design was technically limiting to answer the observed glial specific TRKB.T1 promoter modulation as a functional consequence of methylation alteration with direct experimental evidence. However, the authors attributed possibilities of either astrocyte-specific assorted methylation machinery unique from neurons or astrocyte-specific gene regulatory mechanism different from neurons, which could be responsible for the observed pattern of epigenetic regulation of TRKB.T1 expression. Although the study witnessed serious limitation to provide more mechanistic insights into TRKB promoter methylation regulation for synthesizing TRKB.T1 isoform, it still established a strong association with suicidality in a relatively limited population size (n = 10) free from the effect of confounding variables like PMI, pH, age and substance abuse.
3. Influence of histone modification based epigenetic regulation in suicide pathophysiology: knowledge from post-mortem brain samples
Histones are basic proteins in their chemical constitution and functionally involved in maintaining a three dimensional chromatin conformation by forming nucleosomal core structures in eukaryotic nuclei (Venkatesh and Workman, 2015). The structural framework of nucleosome lies in the formation of octamer unit with dimers of four different histone isoforms H2A, H2B, H3 and H4. Approximately, 147 base pairs of DNA wrapping around the histone octamer with a 1.67 left handed superhelical turn usually connects two adjacent nucleosome cores with an 80bp long linker DNA thread. Dynamic switching of different histone variants and post-translation modification of N-terminal protruded tails (PTM) of core histone molecules results in transient nuclear DNA breathing. This switching helps to acquire either an open (euchromatin) or a closed (heterochromatin) chromatin conformation representative of an active or repressed state of gene transcription (Luger et al., 2012). The N-terminal protruded tails of histones are prone to get various covalent reversible modifications including acetylation, methylation, phosphorylation and many more (Li et al., 2007), but distinct histone modification profiles such as histone 3 lysine 4 (H3K4) dimethylation and trimethylation, and histone 3 lysine 27 (H3K27) acetylation at promoter regions and H3K4 monomethylation in enhancer regions of genes are associated with active gene transcription, whereas repressed promoters are associated with H3K9 and H3K27 dimethylation and trimethylation marks (Turecki, 2014). Collectively, this tail based various histone modifications and associated biochemical modifiers like histone methyltransferase [HMT] and histone deacetylase [HDAC]) build up a potential epigenetic mechanism to transduce the extracellularly received environmental signals into the intracellular milieu with active modulation of gene transcription (Maze et al., 2013).
Reversible histone modification as environmentally induced epigenomic footprint has remained an encouraging field to study in order to understand psychiatric illnesses at the molecular level. A plethora of scientific reports have poured in over last few years to understand the altered histone code in explaining mental disorders such as major depression and schizophrenia (Gavin and Sharma, 2010; Sun et al., 2013; Tsankova et al., 2007), but adequate knowledge is still lacking in interpreting the epigenomic complexities of suicidal behavior from histone modification perspective except for a few interesting reports in both post-mortem samples and in blood cells of suicide attempters.
A. Involvement of neurotrophic receptors
One of the first studies conducted by Ernst and colleagues in 2009 brought attention to certain changes in the histone tails as possible epigenetic modification in post-mortem brains (Ernst et al., 2009). The observed methylation alteration of histone residues was thought to be the reason behind the down regulation of TRKB and TRKB.T1 genes in the brain samples of suicide and major depression cases. Closer examination in the orbital frontal cortex (Brodmann Area 10) of 20 suicide completers revealed a significant increase in type 3 histone (H3) lysine (K) methylation in core octamer of nucleosome close to the TRKB.T1 promoter. The possibility of including any erroneous astrocyte-specific genomic fragments in the chromatin pull-down group was nullified by assaying the promoter enrichment for glutamine synthetase gene. Altogether, the experimental outcome first elucidated the role of H3K27 methylation based repressive chromatin modification in suicide brains with depression background, devoid of any psychotic involvement. Furthermore, a significant inverse correlation was identified between the expression level of TRKB.T1 gene in the same brain area and the observed histone methylation status in its promoter region which necessarily signifies the functional involvement of the repressive epigenetic mark on altered TRKB.T1 expression.
B. Involvement of polyamine system
In the following years, three subsequent studies by Fiori and colleagues in Brodmann areas cited the involvement of histone methylation as key events in regulating the expression of polyamine genes in suicide victims (Fiori and Turecki, 2010, 2011; Fiori and Turecki, 2012). However, two of the dorsolateral prefrontal cortex based studies (Fiori and Turecki, 2010, 2011) in suicide victims were found to be interesting due to the absence of finding any significant changes in the methylation of the 27th lysine residue in the histone-3 tail (H3K27). These two studies were done to identify the repressive role of H3K27 tri-methylation in downregulating the SMS, SMOX, and SAT1 genes by altering their promoter chromatin conformation. Although a functional connection between H3K27me3 and repressed gene expression was found to be lacking in these two studies, it seems to be important for the involvement of other histone-based repressive modifications like H3K9me3 to explain the modulated expression of three aforementioned polyamine genes in suicide brain. Additionally, in-depth consideration are also expected to justify the importance of area specific epigenetic modifications in suicide brains in interpreting the lack of correlation between the histone methylation and corresponding gene expression as observed in these two studies. Most interestingly, the third study from the same group was successful in indicating an association between the H3K4me3 based increased methylation levels with higher risk of suicide (Fiori and Turecki, 2012). The study explained the importance of activating histone-based chromatin modification to transcriptionally induce OAZ1 expression, essentially involved in polyamine biosynthesis and found to be upregulated in the suicide brain. Compelling evidence from this study helps to make a comprehensive understanding of the epigenetically impaired polyamine biosynthesis system in suicide neurobiology. An analysis of 34 suicide brains in BA 44 region identified a significantly elevated level of OAZ1 expression unlike the other three genes (AMD1, ARG2, or OAZ2) examined. Upstream promoter analysis following chromatin immunoprecipitation displayed significantly elevated levels of H3K4me3 in the same 34 suicide completers and this inducible chromatin modification was found to be directly correlated with upregulated OAZ1 expression. The lack of observation of either significant differences in the levels of H3K4me3 in the promoter regions of AMD1, ARG2, or OAZ2, or the expression of AMD1 or OAZ2 leaves the possibility of distinct epigenetic regulation of these.
Collectively, all the evidence discussed above portrays the increasing need of understanding histone-based epigenetic regulation to address the neurobiological complexities associated with suicide. Genome-wide histone modification study could possibly open up and help to explore altered epigenomic regulatory networks as the underlying cause of suicide to interpret the complex nature of this disease or may eventually help to design effective diagnostic strategy for earlier prevention of this mental disorder.
C. Involvement of gap junctional protein connexin
Being primary contributor of glial cell lineage (Kimelberg and Nedergaard, 2010), dysfunctional astrocyte network has been implicated as one of the underlying causes in conferring depressive phenotype (McNally et al., 2008; Rial et al., 2015). Family of connexin genes including CX30 and CX43 (Koulakoff et al., 2008) were found to be heavily impaired in PFC of depressed brain as part of this astrocyte network (Koyama, 2015)which failed to build up proper inter-astrocytic connection with functionally active gap junctional channels (Ernst et al., 2011; Sun et al., 2012). This observation was extended by Nagy and group (Nagy et al., 2016), where a previously reported downregulation of CX30 and CX43 genes in PFC of suicide brain had been shown to be closely associated with epigenetic modifications at their promoter regions (Ernst et al., 2011). The functional contribution of associated epigenetic change on repressing CX gene expression was found to be mediated by a repressive histone modification (H3K9me3) in BA8/9 regions of suicide brain (Nagy et al., 2016). Although a negative correlation was observed between the enriched H3K9me3 modification and gene expression of both CX genes (CX30 and CX43), a significant correlation was only observed for CX30 gene which was found to be free from any confounding effects of variables such as age, tissue pH, or PMI. This histone-specific modification of CX genes provided further insight in explaining the role of alternative epigenetic regulation in causing diminished expression of CX30 and 43 genes which were earlier (Nagy et al., 2015) not supported by site-specific methylation changes at DNA level (CpG based DNA methylation). Moreover, findings from this study also indicated an abrupt change in two CX genes resulting in the loss of correlation between their expressions status, except the subcortical area of 22 suicide brains, where it was found to be tightly correlated in all over neocortex of matched control subjects. Furthermore, an altered cellular morphology of astrocyte has been documented with a corresponding change in CX gene expression downregulation (Olk et al., 2009). The indicated hypertrophic nature of astrocytic population could be due to cytoskeletal deformities arising from CX gene hypofunction. Taken together, this study (Nagy et al., 2016) represented a new dimension of histone mediated epigenetic regulation on cellular morphology and connectivity in moderating PFC function of depressed suicide brain associated with glial lineage.
4. Future directions
A. Epigenetic signature in peripheral blood mononuclear cells-an emerging diagnostic approach to explore suicide biomarker
The recent emergence of peripheral blood mononuclear cells as a reliable source to study and analyze the epigenetic signature of neurological disorders seems to be promising and offers an adequate scope to develop predictive biomarkers (Arosio et al., 2014; Fan et al., 2014; Munkholm et al., 2015; Sharma, 2012). Suicide is a complex process encompassing ideation, planning, attempting, and finally committing suicide (Klonsky et al., 2016). Thus, it would be more logical to study the associated epigenetic changes at every step in suicidal patients, as this will provide ample scope for environmental influence to be worked upon. In this connection, peripheral blood samples would be more accessible and minimally invasive tissue source to carry out biochemical analysis for identifying any corresponding changes (Rao et al., 2013). Emergent application of peripheral blood mononuclear cell transcriptome analysis provides a futuristic surrogate marker concept for assessing neurologic diseases as well as neuropsychiatric disorders (Karsten et al., 2011; Rollins et al., 2010). Moreover, using peripheral blood mononuclear cells (PBMC) over post-mortem brain samples to study epigenetic alteration related to suicide behavior offers an additional advantage of less interfered chemical environment which is relatively challenging in brain samples obtained from suicide victims because of unwanted acidosis.
Considering all these compelling facts, Kang and colleagues in 2013 first reported the successful observation of increased BDNF gene promoter DNA methylation in major depressive patients showing a significant association with previous history of suicidal attempt as well as with suicidal ideation (Kang et al., 2013). This hypermethylated BDNF gene promoter parallels the earlier observation of decreased BDNF gene expression in peripheral blood mononuclear cells of depressed patients with earlier history of attempted suicide (Lee and Kim, 2010). Under the study, the degree of BDNF promoter methylation was found to be correlated with suicidal ideation across 108 patients diagnosed with MDD. The outcome of this PBMC based study strongly suggests the possible use of observed BDNF promoter methylation as proxy epigenetic biomarker for suicidal behavior in depressive patients.
The next piece of study was conducted in East Asian population where prevalence of suicidal rate among elderly adults was found to contribute a major portion in worldwide suicide incidences (Kim et al., 2014). The objective of the study was to find out a possible connection between BDNF promoter methylation with the prevalence and incidence of suicidal ideation in late life. Based on the earlier observation of DNA methylation based BDNF promoter regulation in Wernicke’s area of suicidal brain (Keller et al., 2010), the group postulated to observe similar epigenetic signature in venous blood leukocytes of 93 patients with strong suicidal ideation. Biochemical analysis of a GC-rich region around the vicinity of BDNF transcript identified five prominent sites with significantly hypermethylated status which were strongly associated with the prevalence of suicidal ideation at baseline as well as over a 2 year follow-up period. In conclusion, the study clearly represents the epigenetic modulation of BDNF gene in peripheral blood which could potentially be used in high risk assessment of present suicidal ideation and could be predictive of future suicidal behavior in elderly population.
With the increasing knowledge on the epigenetic involvement of peripheral blood-based biomarker analysis, certain studies have made remarkable contribution in understanding suicide behavior in the last few years (Guintivano et al., 2014; Kang et al., 2013). One of them was a continuation (Kaminsky et al., 2015) of the study of epigenetic profiling of SKA2 gene which was discussed earlier in the postmortem brain section. The same group had taken initiative to predict the biomarker value of a DNA methylation associated polymorphic locus (rs7208505) in SKA2 gene in a study population of 466 individuals following a noninvasive approach of analyzing their peripheral blood tissue (Sadeh et al., 2016). The study was aimed to ascertain the reliability of this specific SNP linked epigenetic signature on SKA2 3′UTR and its association with suicide risk where high degree of mental trauma was involved as a precipitating factor. With an attempt to associate the epigenetic variation at ‘C’ allele of (rs7208505) SKA2 locus (CpG cg13989295) with suicidal thoughts and behaviors, the study was found to be successful in establishing a strong relationship between SKA2 and suicidal behavior in the living cohort of military individuals (Sadeh et al., 2016). This finding happened to be congruent with an earlier report from an independent study cohort of civilians where the subjects experienced major trauma in their lifetime before going through the investigational procedure (Niculescu et al., 2015a; Niculescu et al., 2015b).
Harnessing more predictive information using the blood-based biomarker approach in order to interpret the probability of hospitalizing subjects due to unsuccessful suicidal attempt turned out to be more molecular with few recent studies (Blasco-Fontecilla et al., 2013; Le-Niculescu et al., 2013; Niculescu et al., 2015b). As can be seen from the recent report by Niculescu and group, a peripheral blood based expression profile of certain genes represents a high degree of significance from their active participation to correlate no suicidal ideation to high suicidal ideation state in 37 individuals from a large cohort of 217 participants (Niculescu et al., 2015b). Initial screening of those genes based on their expression variability were later used in a two-step process to sort out the top most candidate genes with higher potential value as molecular biomarker once assessed for their functional relevance to suicidal ideation. The interesting study design behind this report made it possible to validate the involvement of participating gene set in suicidal thought by analyzing their expression status in a cohort of 26 suicide completers (Niculescu et al., 2015b). Overall, the three steps procedure (discovery for ideation, prioritization based on literature evidence, validation for behavior in completers) to select the top ranking molecular markers considering both up and down regulated expression status ultimately resulted in short listing of 8 genes notably SAT1, SKA2, SLC4A4, KIF2C, MBP, IL6, JUN and KLHDC3 in predicting suicidal ideation (the gene set presented here is free of Bonferroni correction). However, identification of SAT1 and SKA2 genes was potentially significant in establishing a close relationship with suicidal ideation because of their previously established role as biomarker for suicidal behavior (Guintivano et al., 2014; Le-Niculescu et al., 2013).
Epigenetic involvement of serotonergic system in predicting suicidal behavior did not receive much attention or rather appeared inconclusive due to week experimental evidences. However, in a recent report Bani-Fatemi and group attempted to find out a functional link between hyposerotonergic neurotransmission and 25 methylated polymorphic loci of HTR2A gene in blood genomic DNA of 32 suicide attempters with major psychosis (Bani-Fatemi et al., 2016). Though the study was unable to establish a possible connection between the 5HT based epigenetic changes and suicide risk in schizophrenia patients but was able to determine significant hypermethylation at a genetically variable locus (rs6313) in the exon I of HTR2A gene. This was in agreement with the previous report which indicated a possible association of suicide attempts with the hypermethylation status of the same polymorphic locus in patients with schizophrenia background (De Luca et al., 2009).
B. Therapeutic intervention as preventive strategy against aberrant epigenetic modulation in suicide behavior
Disease specific epigenetic footprints is a coordinated biochemical process (Tsankova et al., 2007) which could be intervened with empirical pharmacokinetic modeling to develop novel therapeutics (Karahoca and Momparler, 2013; Kelly et al., 2010; Nebbioso et al., 2012; Sun et al., 2013). Recent advancement in pharmacological research has put forward a series of inhibitors with promising results in silencing active epigenetic states related to schizophrenia, depression, and bipolar disorder (Narayan and Dragunow, 2010; Szyf, 2009). Due to an unarguable predominance of psychiatric illnesses in determining the increasing risk of suicidal behavior (Bertolote and Fleischmann, 2002), an overwhelming need to further consolidate the possible involvement of common epigenetic pathways as potential therapeutic targets with proper psychopharmacological intervention is largely unanswered. Furthermore, long standing neurobiological studies have pointed out the role of impaired cognitive functions originating from psychiatric disorders as one of the crucial determinants of the elevated risk of suicidal behavior (Keilp et al., 2001; Van Heeringen and Marusic, 2003). Keeping this in mind, recent understanding of a well-established connection between epigenetic perturbation and cognitive dysfunction (Day and Sweatt, 2012) could potentially serve as the bridge to decide upon formulating novel therapeutic strategies to lower the risk of attempting suicide and related thoughts. However, a comprehensible input is still lacking in devising a proper therapeutic approach to repair the epigenetic defects accrued in course of developing complex suicidal behavior. Though a considerable number of studies have shown the usefulness of antipsychotic and antidepressant drugs like clozapine and lithium in diminishing the suicidal risks (Griffiths et al., 2014), their effective administration in reversing the epigenetic aberrations is yet to be dissected with greater resolution. Pre-clinical studies have shown that chronic administration of imipramine can lead to hyperacetylation of histone3 (H3) to induce transcriptional activation at BDNF promoter, which may have a role in improving cognitive behavior (Tsankova et al., 2006). Similarly, dimethylation of H3K9 and H3K27 by imipramine (Wilkinson et al., 2009) was found to be equally effective in upregulating transcription of genes which could potentially be directed towards reversing dysfunctional cognitive system. Despite the controversy related to its recreational use and abuse, ketamine has been used to stabilize the mood with a moderate dose by augmenting epigenetic alteration in neurochemical pathways with its fast mode of antidepressant function (Choi et al., 2015). Recent evidence has shown its effectiveness in moderating the impulsivity associated with suicidal ideation of mood disorder patients (Price et al., 2014; Thakurta et al., 2012). Likewise, pharmaco-chemical intervention with valproic acid as histone deacetylase inhibitor (HDACi) in stabilizing the mood could prove to be equally effective in mitigating the risk of suicide attempts because of high life time prevalence of affective disorder (30%–40%) among suicidal patients (Hesdorffer et al., 2010; Oquendo et al., 2011). From pre-clinical studies, similar speculation could be advocated for Trichostatin A for their proven role in long-term potentiation to induce histone acetylation and DNA demethylation with increased efficiency in synaptic transmission (Levenson et al., 2004). Falling in the same category with valproic acid, sodium butyrate, a short chain fatty acid compound, has also been shown to modulate H3 acetylation for upregulating expression of neurotrophic factors such as BDNF and GDNF in brain astrocyte population (Wu et al., 2008). In addition to its primary activity as HDACi, sodium butyrate has been found to act on the H3K4 residue in nucleosome complex to induce trimethylation level (Gupta et al., 2010). Apart from targeting histone modifiers, finding novel pharmacological compound to unplug DNA methylation machinery from being recruited to CpG cites on genome could be an effective strategy to restore gene function which is mostly affected in suicidal brain. The compounds such as 5-aza-dC (AZA), zebularine and doxorubicin were earlier reported to minimize the DNA methyltransferase (DNMTs) activity as well as inducing H3 acetylation in the promoter area of CNS related genes like BDNF in the hippocampal region (Grayson and Guidotti, 2013; Kundakovic et al., 2007; Levenson et al., 2006). Therefore, they may hold a great deal of promise to be implicated as DNMT inhibitors (DNMTi) in a dose-dependent manner to reverse the gene function found to be altered in patients suffering from psychiatric disorders with known history of suicidal behavior. While assessing the scope of designing a more effective therapeutic strategy (Table 2), it is pertinent to understand the intrinsic pattern of epigenetic signature associated with this complex disorder.
Table 2.
Future perspective of epiegentic psychophramacology in suicidal behavior
| Drug Type | Compund Name | Target | Function |
|---|---|---|---|
| HDACi | Trichostatin A | HDAC Class I and II | H3K9 and H3K14 hyperacetylation |
| Sodium Butyrate | HDAC Class I and II | H3 acetylation | |
| Suberoylanilide hydroxamic acid (SAHA) | HDAC Class I and II | H3 and H4 acetylation | |
| Entinostat (benzamide histone deacetylase inhibitor) | HDAC Class I | H3 acetylation | |
| Antidepressant | Imipramine | Monoamine transporter | H3 Acetylation and H3K4 dimethylation |
| Fluoxitine | Monoamine transporter | H3 acetylation | |
| Phenelzine, Tranylcypramine | Lysine-specific demethylase 1 (LSD1) | H3K4 methylation | |
| Mood Stabilizer | Valproic Acid | HDAC Class I and II | H3 and H4 acetylation |
| DNMTi | 5-aza-dC (AZA) | DNMT1 | H3 acetylation |
| Doxorubicin | DNMT1 | H3 acetylation | |
| Zebularine | DNMT1 | H3 acetylation |
A strong association of MDD related neuropathology with disrupted neuro molecular circuitry of suicidal brain shares an overlapping pattern of the epigenetic landscape (Autry and Monteggia, 2009). This epigenetic commonality affecting normal gene function could be effectively manipulated with antidepressant drugs having the pharmacological capability to reinstate the pathogenic modifications. In this connection, few prime candidates, notably imipramine, fluoxetine, escitalopram, ketamine, MAOA inhibitors, and 5-azaD could be named for their equal ability to serve as an antidepressant as well as an epigenetic modifier (Boks et al., 2012; Vialou et al., 2013). Support for this statement comes from the preclinical study where a mouse model of stress-induced depression was used (Tsankova et al., 2006). In this study, Bdnf promoter 3 and 4 (responsible for synthesizing transcript variants III and IV) were analyzed to see the corresponding changes in chromatin conformation associated with histone modifications in response to chronic imipramine treatment (Tsankova et al., 2006). Interestingly, a well-designed experiment with a series of chromatin conformation analysis reported a long-lasting, more than twofold hyperacetylation at histone H3 selectively at Bdnf P3 and P4 under chronic imipramine treatment. Histone acetylation of this kind is associated with open chromatin conformation (Felsenfeld and Groudine, 2003) which helps to de-repress the transcription of downstream genes from the promoter. The most important part of this study was the observation of a non-responsive acetylation status at Bdnf P3 and P4 in control (nondefeated) mice with chronic imipramine administration. The underlying causes of these molecular changes showed that an alteration in Hdac5 and Hdac9 mRNA expression from class I and class II Histone Deacetylases (HDACs) were successfully linked with chronic imipramine treatment in the hippocampus of defeated mice. This proved the potential of imipramine to act as epigenetic modifier in addition to its regular pharmacological role as antidepressant (Nghia et al., 2015).
Over the past few years, compelling evidence from studies related to suicidal behavior have pointed out the pharmacological role of ketamine, an anesthetic agent in antagonizing the glutamate N-methyl-d-aspartate (NMDA) receptor (Kashani et al., 2014; Lee et al., 2015; Niciu et al., 2013). Having been tested as an effective antidepressant drug in patients with depression and treatment-resistant depression (TRD), ketamine was found to reduce clinician-rated explicit suicidal cognition 24 hr after its infusion which can last for up to 10 days (Price et al., 2014). While trying to understand the mechanistic changes associated with this fast mode of anti-suicidal response upon ketamine administration, one clinical correlate appeared to be massively impacted i.e. depression (Price and Mathew, 2015). An overall reversal of depressive symptom in the same study cohort found to be paralleled with the observed anti-suicidal tendency which indicated a positive role of ketamine in regulating AMPA to NMDA receptor throughput, changes in synaptic connections and enhancing the BDNF signaling (Price et al., 2014). However, a link was still missing to establish the intermediate role of ketamine as epigenetic modulator until a recent report from preclinical study pointed out HDAC5 modulation in hippocampal neurons (Choi et al., 2015). The report helped to understand ketamine’s role in rapidly stimulating histone deacetylase 5 (HDAC5) phosphorylation and nuclear export in rat hippocampal neurons with direct involvement of calcium/calmodulin kinase II- and protein kinase D-dependent pathways. Taken together, the above reports helped us remarkably to build up a consolidated therapeutic link between the anti-depressive (Reus et al., 2013) and anti-suicidal (Reinstatler and Youssef, 2015; Wilkinson and Sanacora, 2016) property of ketamine which could be potentially mediated by HDAC5 modulation on downstream genes.
C. Future perspective of glial epigenetics in understanding suicide neurobiology
In early 90s, the delineation of ‘tri-partite’ concept of astrocytes (Araque et al., 2014; Araque et al., 1999) and their spatial association with synaptic junctions endowed the researcher with a detailed introspection of glial cell contribution in critically affecting cognitive responses like memory and mood (Kreisel et al., 2014; Pearson-Leary et al., 2015; Rial et al., 2015). Later on, the identification of microglial cell and their dynamic involvement in numerous synaptic junctions by providing cytokines (Rebola et al., 2011), purines (George et al., 2015; Pascual et al., 2012), glutamate and D-serine (Scianni et al., 2013), NO (Zhan et al., 2014) or BDNF (Gomes et al., 2013; Parkhurst et al., 2013) had helped to forward the ‘quad-partite’ theory of glial contribution in understanding the critical role of bidirectional communication between astrocyte and neurons to reshape the plasticity. The possible loss of aforementioned synaptic plasticity under psychiatric disorders have been found to be correlated with functional deficiency in gliotransmitters such as D-serine (Henneberger et al., 2010; Panatier et al., 2006; Yang et al., 2003), glutamate (Fellin et al., 2004), ATP (Koizumi et al., 2003; Pankratov and Lalo, 2015; Zhang et al., 2003) or adenosine (Newman, 2003; Pascual et al., 2005; Serrano et al., 2006), and/or astrocyte-specific neurotransmitter receptors (Kettenmann and Ransom, 2005). This functional understanding of glial cell biology, which is far from its apparent role in metabolically supporting neuronal system, was further re-enforced from a number of studies conducted in postmortem brains of MDD subjects and suicide completers (Ernst et al., 2009; Ernst et al., 2011; Klempan et al., 2009b; Pandey et al., 2012; Sequeira et al., 2009; Torres-Platas et al., 2011). Detailed cellular and biochemical examination interestingly unfolded the abrogated vesicular traffic and inter-membrane transportation system behind the dysfunctional astrocytic population in selective brain regions such as fronto-cortices (Altshuler et al., 2010; Cao et al., 2013; Rajkowska and Stockmeier, 2013). Such astrocytic disabilities in the fronto-limbic region result in compromised plasticity related adaptive functions which are supported by preclinical studies where reversal of compromised astrocytic function has been observed with administration of antidepressants like SSRI and/or fluoxetine (Hertz et al., 2015; Kong et al., 2009). Perhaps, this could be bridged with the established hypothesis of mood disorder related compromised neuroplasticity in suicide brain (Fuchsova et al., 2015) subjected to the variable adaptive response from environmental stimuli. Over the course of time, advancement in molecular techniques and in-depth molecular dissection have put forward several epigenetic mechanisms with regulatory impact on many neural signaling pathways and individual genes but less has been documented in thoroughly examining the associated molecular pathways at glial level. Therefore, considerable attention needs to be paid on this aspect of glial cell biology where the magnitude of DNA methylation, histone modifications or non-coding RNA-mediated epigenetic regulation could be eventually linked with dysregulated astrocytic gene function to understand dysfunctional fronto-limbic axis in suicide brain in response to environmental cues. Fewer studies (as mentioned before in this review) have been conducted to explore the manifold epigenetic impact of the glial cell population in answering compromised neurotrophic factor related signaling pathways using post-mortem brain, but, a lot can be done by deploying the emergent concepts of DNA hydroxymethylation, long-non coding RNA mediated transcriptional regulation or conformational chromatin capture mediate long-distance gene regulation as a future endeavor to bring about more of the intense functional commitment from glial cell population to answer questions of suicide epigenetics.
5. Concluding remarks
Recently, other types of epigenetic players such as hydroxyl methylated DNA and non-coding RNAs have shown promising influence on distinctly regulating gene expression at both transcription and post-transcription level related to several psychiatry disorders (Grayson and Guidotti, 2014; Kocerha et al., 2015), but their role needs to be equally expanded in understanding the pathobiology of suicide behavior. The recent identification of Ten-Eleven-Translocation family protein TET and its involvement in modifying methylated cytosine to hydroxymethylated cytosine residue has gained considerable attention as a potential epigenetic modifier for activating gene expression (Pastor et al., 2013; Tan and Shi, 2012). Variable representation of 5hmC content across different tissues in vertebrates and its relative enrichment (nearly 10 fold higher if compared with embryonic stem cells) in brain (Kato and Iwamoto, 2014; Kinde et al., 2015) makes it a pertinent choice to explore its relevance in regulating higher order chromatin structure in brain regions such as frontal cortex (accounting for 25% over all other GC modifications) (Kinde et al., 2015), which is mostly susceptible to suicide (Furczyk et al., 2013). This pervasive trend of 5hmC based DNA modification in CNS was further found to be functionally accompanied with neural plasticity related phenotypes with pronounced adaptive behavioral features in regulating higher order cognitive functions by modulating genes such as BDNF IX and FGF 1B (Sun et al., 2014). Additional data on TET 3 isoform, known for its functional abundance in both cortex and hippocampus for intrinsically modulating the magnitude of DNA 5hmC modification, was found to be closely associated with homeostatic plasticity (Yu et al., 2015), which enunciates the need to understand its imperative role in compromising synaptic plasticity of suicidal brain. On the other hand, microRNAs, small RNA molecules from non-coding RNA family, have made a remarkable impact in understanding their epigenetic role as post-transcriptional modifier in dysfunctional gene regulatory network in several central nervous systems (CNS) related psychiatric disorders (Dwivedi, 2011, 2013, 2014). Although, an in-depth discussion of these two major epigenetic modifications in relation to suicide is beyond the scope of this review, anticipating their functional relevance and future perspective of other epigenetic reagents like methylated RNA molecules and long non-coding RNAs is important (Aliperti and Donizetti, 2016; Barry et al., 2014; Satterlee et al., 2014; Zuo et al., 2016). Receiving more mechanistic insight to explore the underlying inter-molecular network, acting as a delicate interface to receive environmental inputs found to be vulnerable to the magnitude of external adversities with increasing risk of suicidal phenotype, will be more helpful to devise effective strategies for early assessment of suicidal behavior and prevention of suicide attempt.
Highlights.
Influence on suicidal epigenome by environmental cues
Reversible histone and 5-methyl cytosine based DNA modifications are prime candidates responsible for epigenomic alteration
Psychoepipharmacological intervention as a promising therapeutic tool to improve suicide neuropathology.
Acknowledgments
This research was supported by grants from the National Institute of Mental Health (R01MH082802; R21MH081099; 1R01MH101890; R01MH100616; 1R01MH107183) and American Foundation for Suicide Prevention (SRG-1-042-14) to Dr. Y. Dwivedi.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aliperti V, Donizetti A. Long Non-coding RNA in Neurons: New Players in Early Response to BDNF Stimulation. Frontiers in molecular neuroscience. 2016;9:15. doi: 10.3389/fnmol.2016.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen NJ. Astrocyte regulation of synaptic behavior. Annual review of cell and developmental biology. 2014;30:439–463. doi: 10.1146/annurev-cellbio-100913-013053. [DOI] [PubMed] [Google Scholar]
- Altshuler LL, Abulseoud OA, Foland-Ross L, Bartzokis G, Chang S, Mintz J, Hellemann G, Vinters HV. Amygdala astrocyte reduction in subjects with major depressive disorder but not bipolar disorder. Bipolar Disord. 2010;12:541–549. doi: 10.1111/j.1399-5618.2010.00838.x. [DOI] [PubMed] [Google Scholar]
- Appel K, Schwahn C, Mahler J, Schulz A, Spitzer C, Fenske K, Stender J, Barnow S, John U, Teumer A, Biffar R, Nauck M, Volzke H, Freyberger HJ, Grabe HJ. Moderation of adult depression by a polymorphism in the FKBP5 gene and childhood physical abuse in the general population. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2011;36:1982–1991. doi: 10.1038/npp.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araque A, Carmignoto G, Haydon PG, Oliet SH, Robitaille R, Volterra A. Gliotransmitters travel in time and space. Neuron. 2014;81:728–739. doi: 10.1016/j.neuron.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araque A, Parpura V, Sanzgiri RP, Haydon PG. Tripartite synapses: glia, the unacknowledged partner. Trends in neurosciences. 1999;22:208–215. doi: 10.1016/s0166-2236(98)01349-6. [DOI] [PubMed] [Google Scholar]
- Arnett MG, Muglia LM, Laryea G, Muglia LJ. Genetic Approaches to Hypothalamic-Pituitary-Adrenal Axis Regulation. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2016;41:245–260. doi: 10.1038/npp.2015.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arosio B, D’Addario C, Gussago C, Casati M, Tedone E, Ferri E, Nicolini P, Rossi PD, Maccarrone M, Mari D. Peripheral blood mononuclear cells as a laboratory to study dementia in the elderly. BioMed research international. 2014;2014:169203. doi: 10.1155/2014/169203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autry AE, Monteggia LM. Epigenetics in suicide and depression. Biological psychiatry. 2009;66:812–813. doi: 10.1016/j.biopsych.2009.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autry AE, Monteggia LM. Brain-derived neurotrophic factor and neuropsychiatric disorders. Pharmacological reviews. 2012;64:238–258. doi: 10.1124/pr.111.005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bani-Fatemi A, Howe AS, De Luca V. Epigenetic studies of suicidal behavior. Neurocase. 2015;21:134–143. doi: 10.1080/13554794.2013.826679. [DOI] [PubMed] [Google Scholar]
- Bani-Fatemi A, Howe AS, Matmari M, Koga A, Zai C, Strauss J, De Luca V. Interaction between Methylation and CpG Single-Nucleotide Polymorphisms in the HTR2A Gene: Association Analysis with Suicide Attempt in Schizophrenia. Neuropsychobiology. 2016;73:10–15. doi: 10.1159/000441191. [DOI] [PubMed] [Google Scholar]
- Barry G, Briggs JA, Vanichkina DP, Poth EM, Beveridge NJ, Ratnu VS, Nayler SP, Nones K, Hu J, Bredy TW, Nakagawa S, Rigo F, Taft RJ, Cairns MJ, Blackshaw S, Wolvetang EJ, Mattick JS. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Molecular psychiatry. 2014;19:486–494. doi: 10.1038/mp.2013.45. [DOI] [PubMed] [Google Scholar]
- Bertolote JM, Fleischmann A. Suicide and psychiatric diagnosis: a worldwide perspective. World psychiatry: official journal of the World Psychiatric Association (WPA) 2002;1:181–185. [PMC free article] [PubMed] [Google Scholar]
- Binder EB. The role of FKBP5, a co-chaperone of the glucocorticoid receptor in the pathogenesis and therapy of affective and anxiety disorders. Psychoneuroendocrinology. 2009;34(Suppl 1):S186–195. doi: 10.1016/j.psyneuen.2009.05.021. [DOI] [PubMed] [Google Scholar]
- Binder EB, Bradley RG, Liu W, Epstein MP, Deveau TC, Mercer KB, Tang Y, Gillespie CF, Heim CM, Nemeroff CB, Schwartz AC, Cubells JF, Ressler KJ. Association of FKBP5 polymorphisms and childhood abuse with risk of posttraumatic stress disorder symptoms in adults. Jama. 2008;299:1291–1305. doi: 10.1001/jama.299.11.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder EB, Salyakina D, Lichtner P, Wochnik GM, Ising M, Putz B, Papiol S, Seaman S, Lucae S, Kohli MA, Nickel T, Kunzel HE, Fuchs B, Majer M, Pfennig A, Kern N, Brunner J, Modell S, Baghai T, Deiml T, Zill P, Bondy B, Rupprecht R, Messer T, Kohnlein O, Dabitz H, Bruckl T, Muller N, Pfister H, Lieb R, Mueller JC, Lohmussaar E, Strom TM, Bettecken T, Meitinger T, Uhr M, Rein T, Holsboer F, Muller-Myhsok B. Polymorphisms in FKBP5 are associated with increased recurrence of depressive episodes and rapid response to antidepressant treatment. Nature genetics. 2004;36:1319–1325. doi: 10.1038/ng1479. [DOI] [PubMed] [Google Scholar]
- Blasco-Fontecilla H, Lopez-Castroman J, Giner L, Baca-Garcia E, Oquendo MA. Predicting suicidal behavior: are we really that far along? Comment on “Discovery and validation of blood biomarkers for suicidality”. Current psychiatry reports. 2013;15:424. doi: 10.1007/s11920-013-0424-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockbrader K, Feng Y. Essential function, sophisticated regulation and pathological impact of the selective RNA-binding protein QKI in CNS myelin development. Future neurology. 2008;3:655–668. doi: 10.2217/14796708.3.6.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boks MP, de Jong NM, Kas MJ, Vinkers CH, Fernandes C, Kahn RS, Mill J, Ophoff RA. Current status and future prospects for epigenetic psychopharmacology. Epigenetics. 2012;7:20–28. doi: 10.4161/epi.7.1.18688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand-Schieber E, Lowery SL, Werner P. Select ionotropic glutamate AMPA/kainate receptors are expressed at the astrocyte-vessel interface. Brain research. 2004;1007:178–182. doi: 10.1016/j.brainres.2003.12.051. [DOI] [PubMed] [Google Scholar]
- Brent D, Melhem N, Ferrell R, Emslie G, Wagner KD, Ryan N, Vitiello B, Birmaher B, Mayes T, Zelazny J, Onorato M, Devlin B, Clarke G, DeBar L, Keller M. Association of FKBP5 polymorphisms with suicidal events in the Treatment of Resistant Depression in Adolescents (TORDIA) study. The American journal of psychiatry. 2010;167:190–197. doi: 10.1176/appi.ajp.2009.09040576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent DA, Melhem N. Familial transmission of suicidal behavior. The Psychiatric clinics of North America. 2008;31:157–177. doi: 10.1016/j.psc.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Li LP, Wang Q, Wu Q, Hu HH, Zhang M, Fang YY, Zhang J, Li SJ, Xiong WC, Yan HC, Gao YB, Liu JH, Li XW, Sun LR, Zeng YN, Zhu XH, Gao TM. Astrocyte-derived ATP modulates depressive-like behaviors. Nature medicine. 2013;19:773–777. doi: 10.1038/nm.3162. [DOI] [PubMed] [Google Scholar]
- Carballo JJ, Akamnonu CP, Oquendo MA. Neurobiology of suicidal behavior. An integration of biological and clinical findings. Archives of suicide research: official journal of the International Academy for Suicide Research. 2008;12:93–110. doi: 10.1080/13811110701857004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carim-Todd L, Bath KG, Fulgenzi G, Yanpallewar S, Jing D, Barrick CA, Becker J, Buckley H, Dorsey SG, Lee FS, Tessarollo L. Endogenous truncated TrkB.T1 receptor regulates neuronal complexity and TrkB kinase receptor function in vivo. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2009;29:678–685. doi: 10.1523/JNEUROSCI.5060-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC. Understanding suicide: Fact sheet. 2015. [Google Scholar]
- Choi JK, Bae JB, Lyu J, Kim TY, Kim YJ. Nucleosome deposition and DNA methylation at coding region boundaries. Genome biology. 2009;10:R89. doi: 10.1186/gb-2009-10-9-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi M, Lee SH, Wang SE, Ko SY, Song M, Choi JS, Kim YS, Duman RS, Son H. Ketamine produces antidepressant-like effects through phosphorylation-dependent nuclear export of histone deacetylase 5 (HDAC5) in rats. Proc Natl Acad Sci U S A. 2015;112:15755–15760. doi: 10.1073/pnas.1513913112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Cory S, Kidane AH, Shirkey NJ, Marshak S. Brain-derived neurotrophic factor and the development of structural neuronal connectivity. Developmental neurobiology. 2010;70:271–288. doi: 10.1002/dneu.20774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currier D, Mann JJ. Stress, genes and the biology of suicidal behavior. The Psychiatric clinics of North America. 2008;31:247–269. doi: 10.1016/j.psc.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD. Epigenetic treatments for cognitive impairments. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2012;37:247–260. doi: 10.1038/npp.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca V, Viggiano E, Dhoot R, Kennedy JL, Wong AH. Methylation and QTDT analysis of the 5-HT2A receptor 102C allele: analysis of suicidality in major psychosis. Journal of psychiatric research. 2009;43:532–537. doi: 10.1016/j.jpsychires.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Ding Y, Lawrence N, Olie E, Cyprien F, le Bars E, Bonafe A, Phillips ML, Courtet P, Jollant F. Prefrontal cortex markers of suicidal vulnerability in mood disorders: a model-based structural neuroimaging study with a translational perspective. Translational psychiatry. 2015;5:e516. doi: 10.1038/tp.2015.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi Y. Brain-derived neurotrophic factor: role in depression and suicide. Neuropsychiatric disease and treatment. 2009;5:433–449. doi: 10.2147/ndt.s5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi Y. Brain-derived neurotrophic factor and suicide pathogenesis. Annals of medicine. 2010;42:87–96. doi: 10.3109/07853890903485730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi Y. Evidence demonstrating role of microRNAs in the etiopathology of major depression. Journal of chemical neuroanatomy. 2011;42:142–156. doi: 10.1016/j.jchemneu.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi Y. microRNAs as Biomarker in Depression Pathogenesis. Annals of psychiatry and mental health. 2013;1:1003. [PMC free article] [PubMed] [Google Scholar]
- Dwivedi Y. Emerging role of microRNAs in major depressive disorder: diagnosis and therapeutic implications. Dialogues in clinical neuroscience. 2014;16:43–61. doi: 10.31887/DCNS.2014.16.1/ydwivedi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi Y, Rizavi HS, Conley RR, Roberts RC, Tamminga CA, Pandey GN. Altered gene expression of brain-derived neurotrophic factor and receptor tyrosine kinase B in postmortem brain of suicide subjects. Archives of general psychiatry. 2003;60:804–815. doi: 10.1001/archpsyc.60.8.804. [DOI] [PubMed] [Google Scholar]
- Ernst C, Deleva V, Deng X, Sequeira A, Pomarenski A, Klempan T, Ernst N, Quirion R, Gratton A, Szyf M, Turecki G. Alternative splicing, methylation state, and expression profile of tropomyosin-related kinase B in the frontal cortex of suicide completers. Archives of general psychiatry. 2009;66:22–32. doi: 10.1001/archpsyc.66.1.22. [DOI] [PubMed] [Google Scholar]
- Ernst C, McGowan PO, Deleva V, Meaney MJ, Szyf M, Turecki G. The effects of pH on DNA methylation state: In vitro and post-mortem brain studies. Journal of neuroscience methods. 2008;174:123–125. doi: 10.1016/j.jneumeth.2008.06.027. [DOI] [PubMed] [Google Scholar]
- Ernst C, Nagy C, Kim S, Yang JP, Deng X, Hellstrom IC, Choi KH, Gershenfeld H, Meaney MJ, Turecki G. Dysfunction of astrocyte connexins 30 and 43 in dorsal lateral prefrontal cortex of suicide completers. Biological psychiatry. 2011;70:312–319. doi: 10.1016/j.biopsych.2011.03.038. [DOI] [PubMed] [Google Scholar]
- Ewald ER, Wand GS, Seifuddin F, Yang X, Tamashiro KL, Potash JB, Zandi P, Lee RS. Alterations in DNA methylation of Fkbp5 as a determinant of blood-brain correlation of glucocorticoid exposure. Psychoneuroendocrinology. 2014;44:112–122. doi: 10.1016/j.psyneuen.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan HM, Sun XY, Guo W, Zhong AF, Niu W, Zhao L, Dai YH, Guo ZM, Zhang LY, Lu J. Differential expression of microRNA in peripheral blood mononuclear cells as specific biomarker for major depressive disorder patients. Journal of psychiatric research. 2014;59:45–52. doi: 10.1016/j.jpsychires.2014.08.007. [DOI] [PubMed] [Google Scholar]
- Faravelli C, Lo Sauro C, Godini L, Lelli L, Benni L, Pietrini F, Lazzeretti L, Talamba GA, Fioravanti G, Ricca V. Childhood stressful events, HPA axis and anxiety disorders. World journal of psychiatry. 2012a;2:13–25. doi: 10.5498/wjp.v2.i1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faravelli C, Lo Sauro C, Lelli L, Pietrini F, Lazzeretti L, Godini L, Benni L, Fioravanti G, Talamba GA, Castellini G, Ricca V. The role of life events and HPA axis in anxiety disorders: a review. Current pharmaceutical design. 2012b;18:5663–5674. doi: 10.2174/138161212803530907. [DOI] [PubMed] [Google Scholar]
- Fellin T, Pascual O, Gobbo S, Pozzan T, Haydon PG, Carmignoto G. Neuronal synchrony mediated by astrocytic glutamate through activation of extrasynaptic NMDA receptors. Neuron. 2004;43:729–743. doi: 10.1016/j.neuron.2004.08.011. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- Fields RD. White matter in learning, cognition and psychiatric disorders. Trends in neurosciences. 2008;31:361–370. doi: 10.1016/j.tins.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiori LM, Bureau A, Labbe A, Croteau J, Noel S, Merette C, Turecki G. Global gene expression profiling of the polyamine system in suicide completers. The international journal of neuropsychopharmacology/official scientific journal of the Collegium Internationale Neuropsychopharmacologicum (CINP) 2011;14:595–605. doi: 10.1017/S1461145710001574. [DOI] [PubMed] [Google Scholar]
- Fiori LM, Turecki G. Implication of the polyamine system in mental disorders. Journal of psychiatry & neuroscience: JPN. 2008;33:102–110. [PMC free article] [PubMed] [Google Scholar]
- Fiori LM, Turecki G. Association of the SAT1 in/del polymorphism with suicide completion. American journal of medical genetics Part B, Neuropsychiatric genetics: the official publication of the International Society of Psychiatric Genetics. 2010;153b:825–829. doi: 10.1002/ajmg.b.31040. [DOI] [PubMed] [Google Scholar]
- Fiori LM, Turecki G. Epigenetic regulation of spermidine/spermine N1-acetyltransferase (SAT1) in suicide. Journal of psychiatric research. 2011;45:1229–1235. doi: 10.1016/j.jpsychires.2011.03.015. [DOI] [PubMed] [Google Scholar]
- Fiori LM, Turecki G. Approaches and Findings from Gene Expression Profiling Studies of Suicide. In: dwivedi Y, editor. The Neurobiological Basis of Suicide. Frontiers in Neuroscience. CRC Press/Taylor & Francis; Boca Raton (FL): 2012. [PubMed] [Google Scholar]
- Fuchsova B, Alvarez Julia A, Rizavi HS, Frasch AC, Pandey GN. Altered expression of neuroplasticity-related genes in the brain of depressed suicides. Neuroscience. 2015;299:1–17. doi: 10.1016/j.neuroscience.2015.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furczyk K, Schutova B, Michel TM, Thome J, Buttner A. The neurobiology of suicide - A Review of post-mortem studies. Journal of molecular psychiatry. 2013;1:2. doi: 10.1186/2049-9256-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galfalvy H, Haghighi F, Hodgkinson C, Goldman D, Oquendo MA, Burke A, Huang YY, Giegling I, Rujescu D, Bureau A, Turecki G, Mann JJ. A genome-wide association study of suicidal behavior. American journal of medical genetics Part B, Neuropsychiatric genetics: the official publication of the International Society of Psychiatric Genetics. 2015;168:557–563. doi: 10.1002/ajmg.b.32330. [DOI] [PubMed] [Google Scholar]
- Gavin DP, Sharma RP. Histone modifications, DNA methylation, and schizophrenia. Neuroscience and biobehavioral reviews. 2010;34:882–888. doi: 10.1016/j.neubiorev.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George J, Goncalves FQ, Cristovao G, Rodrigues L, Meyer Fernandes JR, Goncalves T, Cunha RA, Gomes CA. Different danger signals differently impact on microglial proliferation through alterations of ATP release and extracellular metabolism. Glia. 2015;63:1636–1645. doi: 10.1002/glia.22833. [DOI] [PubMed] [Google Scholar]
- Gerner EW, Meyskens FL., Jr Polyamines and cancer: old molecules, new understanding. Nature reviews Cancer. 2004;4:781–792. doi: 10.1038/nrc1454. [DOI] [PubMed] [Google Scholar]
- Gomes C, Ferreira R, George J, Sanches R, Rodrigues DI, Goncalves N, Cunha RA. Activation of microglial cells triggers a release of brain-derived neurotrophic factor (BDNF) inducing their proliferation in an adenosine A2A receptor-dependent manner: A2A receptor blockade prevents BDNF release and proliferation of microglia. Journal of neuroinflammation. 2013;10:16. doi: 10.1186/1742-2094-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson DR, Guidotti A. The dynamics of DNA methylation in schizophrenia and related psychiatric disorders. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2013;38:138–166. doi: 10.1038/npp.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson DR, Guidotti A. Epigenetics in Psychiatry5-Methycytosine and 5-Hydroxymethylcytosine in Psychiatric Epigenetics, Epigenetics in Psychiatry. Academic press; Tokyo: 2014. pp. 209–240. [Google Scholar]
- Griffiths JJ, Zarate CA, Jr, Rasimas JJ. Existing and novel biological therapeutics in suicide prevention. American journal of preventive medicine. 2014;47:S195–203. doi: 10.1016/j.amepre.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JA, Fiori LM, Labonte B, Lopez JP, Turecki G. Effects of promoter methylation on increased expression of polyamine biosynthetic genes in suicide. Journal of psychiatric research. 2013;47:513–519. doi: 10.1016/j.jpsychires.2012.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti G, Calabrese F, Anacker C, Racagni G, Pariante CM, Riva MA. Glucocorticoid receptor and FKBP5 expression is altered following exposure to chronic stress: modulation by antidepressant treatment. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2013;38:616–627. doi: 10.1038/npp.2012.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guintivano J, Brown T, Newcomer A, Jones M, Cox O, Maher BS, Eaton WW, Payne JL, Wilcox HC, Kaminsky ZA. Identification and replication of a combined epigenetic and genetic biomarker predicting suicide and suicidal behaviors. The American journal of psychiatry. 2014;171:1287–1296. doi: 10.1176/appi.ajp.2014.14010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guipponi M, Deutsch S, Kohler K, Perroud N, Le Gal F, Vessaz M, Laforge T, Petit B, Jollant F, Guillaume S, Baud P, Courtet P, La Harpe R, Malafosse A. Genetic and epigenetic analysis of SSAT gene dysregulation in suicidal behavior. American journal of medical genetics Part B, Neuropsychiatric genetics: the official publication of the International Society of Psychiatric Genetics. 2009;150b:799–807. doi: 10.1002/ajmg.b.30901. [DOI] [PubMed] [Google Scholar]
- Gupta S, Kim SY, Artis S, Molfese DL, Schumacher A, Sweatt JD, Paylor RE, Lubin FD. Histone methylation regulates memory formation. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2010;30:3589–3599. doi: 10.1523/JNEUROSCI.3732-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghighi F, Xin Y, Chanrion B, O’Donnell AH, Ge Y, Dwork AJ, Arango V, Mann JJ. Increased DNA methylation in the suicide brain. Dialogues in clinical neuroscience. 2014;16:430–438. doi: 10.31887/DCNS.2014.16.3/jmann. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanisch A, Sillje HH, Nigg EA. Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. The EMBO journal. 2006;25:5504–5515. doi: 10.1038/sj.emboj.7601426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EC, Barraclough B. Suicide as an outcome for mental disorders. A meta-analysis. The British journal of psychiatry: the journal of mental science. 1997;170:205–228. doi: 10.1192/bjp.170.3.205. [DOI] [PubMed] [Google Scholar]
- Hellstrom IC, Dhir SK, Diorio JC, Meaney MJ. Maternal licking regulates hippocampal glucocorticoid receptor transcription through a thyroid hormone-serotonin-NGFI-A signalling cascade. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2012;367:2495–2510. doi: 10.1098/rstb.2012.0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henneberger C, Papouin T, Oliet SH, Rusakov DA. Long-term potentiation depends on release of D-serine from astrocytes. Nature. 2010;463:232–236. doi: 10.1038/nature08673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JP, Ostrander MM, Mueller NK, Figueiredo H. Limbic system mechanisms of stress regulation: hypothalamo-pituitary-adrenocortical axis. Progress in neuro-psychopharmacology & biological psychiatry. 2005;29:1201–1213. doi: 10.1016/j.pnpbp.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Hertz L, Rothman DL, Li B, Peng L. Chronic SSRI stimulation of astrocytic 5-HT2B receptors change multiple gene expressions/editings and metabolism of glutamate, glucose and glycogen: a potential paradigm shift. Frontiers in behavioral neuroscience. 2015;9:25. doi: 10.3389/fnbeh.2015.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz L, Song D, Li B, Du T, Xu J, Gu L, Chen Y, Peng L. Signal Transduction in Astrocytes during Chronic or Acute Treatment with Drugs (SSRIs, Antibipolar Drugs, GABA-ergic Drugs, and Benzodiazepines) Ameliorating Mood Disorders. Journal of signal transduction. 2014;2014:593934. doi: 10.1155/2014/593934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesdorffer DC, Berg AT, Kanner AM. An update on antiepileptic drugs and suicide: are there definitive answers yet? Epilepsy currents/American Epilepsy Society. 2010;10:137–145. doi: 10.1111/j.1535-7511.2010.01382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoertel N, Franco S, Wall MM, Oquendo MA, Kerridge BT, Limosin F, Blanco C. Mental disorders and risk of suicide attempt: a national prospective study. Molecular psychiatry. 2015;20:718–726. doi: 10.1038/mp.2015.19. [DOI] [PubMed] [Google Scholar]
- Hohne N, Poidinger M, Merz F, Pfister H, Bruckl T, Zimmermann P, Uhr M, Holsboer F, Ising M. FKBP5 genotype-dependent DNA methylation and mRNA regulation after psychosocial stress in remitted depression and healthy controls. The international journal of neuropsychopharmacology/official scientific journal of the Collegium Internationale Neuropsychopharmacologicum (CINP) 2015:18. doi: 10.1093/ijnp/pyu087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoptman MJ, Volavka J, Johnson G, Weiss E, Bilder RM, Lim KO. Frontal white matter microstructure, aggression, and impulsivity in men with schizophrenia: a preliminary study. Biological psychiatry. 2002;52:9–14. doi: 10.1016/s0006-3223(02)01311-2. [DOI] [PubMed] [Google Scholar]
- Jankord R, Herman JP. Limbic regulation of hypothalamo-pituitary-adrenocortical function during acute and chronic stress. Annals of the New York Academy of Sciences. 2008;1148:64–73. doi: 10.1196/annals.1410.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jollant F, Bellivier F, Leboyer M, Astruc B, Torres S, Verdier R, Castelnau D, Malafosse A, Courtet P. Impaired decision making in suicide attempters. The American journal of psychiatry. 2005;162:304–310. doi: 10.1176/appi.ajp.162.2.304. [DOI] [PubMed] [Google Scholar]
- Jollant F, Guillaume S, Jaussent I, Castelnau D, Malafosse A, Courtet P. Impaired decision-making in suicide attempters may increase the risk of problems in affective relationships. J Affect Disord. 2007;99:59–62. doi: 10.1016/j.jad.2006.07.022. [DOI] [PubMed] [Google Scholar]
- Jollant F, Lawrence NS, Olie E, O’Daly O, Malafosse A, Courtet P, Phillips ML. Decreased activation of lateral orbitofrontal cortex during risky choices under uncertainty is associated with disadvantageous decision-making and suicidal behavior. NeuroImage. 2010;51:1275–1281. doi: 10.1016/j.neuroimage.2010.03.027. [DOI] [PubMed] [Google Scholar]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nature reviews Genetics. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Kallman FJ. Heredity in Health and Mental Disease: Principles of Psychiatric Genetics in the Light of Comparative Twin Studies. Norton; New York: 1953. [Google Scholar]
- Kamali M, Oquendo MA, Mann JJ. Understanding the neurobiology of suicidal behavior. Depression and anxiety. 2001;14:164–176. doi: 10.1002/da.1062. [DOI] [PubMed] [Google Scholar]
- Kaminsky Z, Wilcox HC, Eaton WW, Van Eck K, Kilaru V, Jovanovic T, Klengel T, Bradley B, Binder EB, Ressler KJ, Smith AK. Epigenetic and genetic variation at SKA2 predict suicidal behavior and post-traumatic stress disorder. Translational psychiatry. 2015;5:e627. doi: 10.1038/tp.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Kim JM, Lee JY, Kim SY, Bae KY, Kim SW, Shin IS, Kim HR, Shin MG, Yoon JS. BDNF promoter methylation and suicidal behavior in depressive patients. J Affect Disord. 2013;151:679–685. doi: 10.1016/j.jad.2013.08.001. [DOI] [PubMed] [Google Scholar]
- Karahoca M, Momparler RL. Pharmacokinetic and pharmacodynamic analysis of 5-aza-2′-deoxycytidine (decitabine) in the design of its dose-schedule for cancer therapy. Clinical epigenetics. 2013;5:3. doi: 10.1186/1868-7083-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karege F, Vaudan G, Schwald M, Perroud N, La Harpe R. Neurotrophin levels in postmortem brains of suicide victims and the effects of antemortem diagnosis and psychotropic drugs. Brain research Molecular brain research. 2005;136:29–37. doi: 10.1016/j.molbrainres.2004.12.020. [DOI] [PubMed] [Google Scholar]
- Karsten SL, Kudo LC, Bragin AJ. Use of peripheral blood transcriptome biomarkers for epilepsy prediction. Neuroscience letters. 2011;497:213–217. doi: 10.1016/j.neulet.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashani P, Yousefian S, Amini A, Heidari K, Younesian S, Hatamabadi HR. The Effect of Intravenous Ketamine in Suicidal Ideation of Emergency Department Patients. Emergency (Tehran, Iran) 2014;2:36–39. [PMC free article] [PubMed] [Google Scholar]
- Kato T, Iwamoto K. Comprehensive DNA methylation and hydroxymethylation analysis in the human brain and its implication in mental disorders. Neuropharmacology. 2014;80:133–139. doi: 10.1016/j.neuropharm.2013.12.019. [DOI] [PubMed] [Google Scholar]
- Kawamoto EM, Vivar C, Camandola S. Physiology and pathology of calcium signaling in the brain. Frontiers in pharmacology. 2012;3:61. doi: 10.3389/fphar.2012.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keilp JG, Sackeim HA, Brodsky BS, Oquendo MA, Malone KM, Mann JJ. Neuropsychological dysfunction in depressed suicide attempters. The American journal of psychiatry. 2001;158:735–741. doi: 10.1176/appi.ajp.158.5.735. [DOI] [PubMed] [Google Scholar]
- Keller S, Sarchiapone M, Zarrilli F, Tomaiuolo R, Carli V, Angrisano T, Videtic A, Amato F, Pero R, di Giannantonio M, Iosue M, Lembo F, Castaldo G, Chiariotti L. TrkB gene expression and DNA methylation state in Wernicke area does not associate with suicidal behavior. J Affect Disord. 2011;135:400–404. doi: 10.1016/j.jad.2011.07.003. [DOI] [PubMed] [Google Scholar]
- Keller S, Sarchiapone M, Zarrilli F, Videtic A, Ferraro A, Carli V, Sacchetti S, Lembo F, Angiolillo A, Jovanovic N, Pisanti F, Tomaiuolo R, Monticelli A, Balazic J, Roy A, Marusic A, Cocozza S, Fusco A, Bruni CB, Castaldo G, Chiariotti L. Increased BDNF promoter methylation in the Wernicke area of suicide subjects. Archives of general psychiatry. 2010;67:258–267. doi: 10.1001/archgenpsychiatry.2010.9. [DOI] [PubMed] [Google Scholar]
- Kelly TK, De Carvalho DD, Jones PA. Epigenetic modifications as therapeutic targets. Nature biotechnology. 2010;28:1069–1078. doi: 10.1038/nbt.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenmann H, Ransom BR. Neuroglia. 3. Oxford University Press; Oxford: 2005. [Google Scholar]
- Kim CD, Seguin M, Therrien N, Riopel G, Chawky N, Lesage AD, Turecki G. Familial aggregation of suicidal behavior: a family study of male suicide completers from the general population. The American journal of psychiatry. 2005;162:1017–1019. doi: 10.1176/appi.ajp.162.5.1017. [DOI] [PubMed] [Google Scholar]
- Kim JM, Kang HJ, Bae KY, Kim SW, Shin IS, Kim HR, Shin MG, Yoon JS. Association of BDNF promoter methylation and genotype with suicidal ideation in elderly Koreans. The American journal of geriatric psychiatry: official journal of the American Association for Geriatric Psychiatry. 2014;22:989–996. doi: 10.1016/j.jagp.2014.02.011. [DOI] [PubMed] [Google Scholar]
- Kimelberg HK, Nedergaard M. Functions of astrocytes and their potential as therapeutic targets. Neurotherapeutics: the journal of the American Society for Experimental NeuroTherapeutics. 2010;7:338–353. doi: 10.1016/j.nurt.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinde B, Gabel HW, Gilbert CS, Griffith EC, Greenberg ME. Reading the unique DNA methylation landscape of the brain: Non-CpG methylation, hydroxymethylation, and MeCP2. Proc Natl Acad Sci U S A. 2015;112:6800–6806. doi: 10.1073/pnas.1411269112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klempan TA, Rujescu D, Merette C, Himmelman C, Sequeira A, Canetti L, Fiori LM, Schneider B, Bureau A, Turecki G. Profiling brain expression of the spermidine/spermine N1-acetyltransferase 1 (SAT1) gene in suicide. American journal of medical genetics Part B, Neuropsychiatric genetics: the official publication of the International Society of Psychiatric Genetics. 2009a;150b:934–943. doi: 10.1002/ajmg.b.30920. [DOI] [PubMed] [Google Scholar]
- Klempan TA, Sequeira A, Canetti L, Lalovic A, Ernst C, ffrench-Mullen J, Turecki G. Altered expression of genes involved in ATP biosynthesis and GABAergic neurotransmission in the ventral prefrontal cortex of suicides with and without major depression. Molecular psychiatry. 2009b;14:175–189. doi: 10.1038/sj.mp.4002110. [DOI] [PubMed] [Google Scholar]
- Klengel T, Mehta D, Anacker C, Rex-Haffner M, Pruessner JC, Pariante CM, Pace TW, Mercer KB, Mayberg HS, Bradley B, Nemeroff CB, Holsboer F, Heim CM, Ressler KJ, Rein T, Binder EB. Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nature neuroscience. 2013;16:33–41. doi: 10.1038/nn.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klonsky ED, May AM, Saffer BY. Suicide, Suicide Attempts, and Suicidal Ideation. Annual review of clinical psychology. 2016;12:307–330. doi: 10.1146/annurev-clinpsy-021815-093204. [DOI] [PubMed] [Google Scholar]
- Kocerha J, Dwivedi Y, Brennand KJ. Noncoding RNAs and neurobehavioral mechanisms in psychiatric disease. Molecular psychiatry. 2015;20:677–684. doi: 10.1038/mp.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi S, Fujishita K, Tsuda M, Shigemoto-Mogami Y, Inoue K. Dynamic inhibition of excitatory synaptic transmission by astrocyte-derived ATP in hippocampal cultures. Proc Natl Acad Sci U S A. 2003;100:11023–11028. doi: 10.1073/pnas.1834448100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong H, Sha LL, Fan Y, Xiao M, Ding JH, Wu J, Hu G. Requirement of AQP4 for antidepressive efficiency of fluoxetine: implication in adult hippocampal neurogenesis. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2009;34:1263–1276. doi: 10.1038/npp.2008.185. [DOI] [PubMed] [Google Scholar]
- Koulakoff A, Ezan P, Giaume C. Neurons control the expression of connexin 30 and connexin 43 in mouse cortical astrocytes. Glia. 2008;56:1299–1311. doi: 10.1002/glia.20698. [DOI] [PubMed] [Google Scholar]
- Koyama Y. Functional alterations of astrocytes in mental disorders: pharmacological significance as a drug target. Frontiers in cellular neuroscience. 2015;9:261. doi: 10.3389/fncel.2015.00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreisel T, Frank MG, Licht T, Reshef R, Ben-Menachem-Zidon O, Baratta MV, Maier SF, Yirmiya R. Dynamic microglial alterations underlie stress-induced depressive-like behavior and suppressed neurogenesis. Molecular psychiatry. 2014;19:699–709. doi: 10.1038/mp.2013.155. [DOI] [PubMed] [Google Scholar]
- Kubota T, Miyake K, Hirasawa T. Epigenetic understanding of gene-environment interactions in psychiatric disorders: a new concept of clinical genetics. Clinical epigenetics. 2012;4:1. doi: 10.1186/1868-7083-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundakovic M, Champagne FA. Early-life experience, epigenetics, and the developing brain. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2015;40:141–153. doi: 10.1038/npp.2014.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundakovic M, Chen Y, Costa E, Grayson DR. DNA methyltransferase inhibitors coordinately induce expression of the human reelin and glutamic acid decarboxylase 67 genes. Molecular pharmacology. 2007;71:644–653. doi: 10.1124/mol.106.030635. [DOI] [PubMed] [Google Scholar]
- Labonte B, Suderman M, Maussion G, Lopez JP, Navarro-Sanchez L, Yerko V, Mechawar N, Szyf M, Meaney MJ, Turecki G. Genome-wide methylation changes in the brains of suicide completers. The American journal of psychiatry. 2013;170:511–520. doi: 10.1176/appi.ajp.2012.12050627. [DOI] [PubMed] [Google Scholar]
- Labonte B, Suderman M, Maussion G, Navaro L, Yerko V, Mahar I, Bureau A, Mechawar N, Szyf M, Meaney MJ, Turecki G. Genome-wide epigenetic regulation by early-life trauma. Archives of general psychiatry. 2012;69:722–731. doi: 10.1001/archgenpsychiatry.2011.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonte B, Turecki G. The epigenetics of suicide: explaining the biological effects of early life environmental adversity. Archives of suicide research: official journal of the International Academy for Suicide Research. 2010;14:291–310. doi: 10.1080/13811118.2010.524025. [DOI] [PubMed] [Google Scholar]
- Le-Niculescu H, Levey DF, Ayalew M, Palmer L, Gavrin LM, Jain N, Winiger E, Bhosrekar S, Shankar G, Radel M, Bellanger E, Duckworth H, Olesek K, Vergo J, Schweitzer R, Yard M, Ballew A, Shekhar A, Sandusky GE, Schork NJ, Kurian SM, Salomon DR, Niculescu AB., 3rd Discovery and validation of blood biomarkers for suicidality. Molecular psychiatry. 2013;18:1249–1264. doi: 10.1038/mp.2013.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Kim YK. BDNF mRNA expression of peripheral blood mononuclear cells was decreased in depressive patients who had or had not recently attempted suicide. J Affect Disord. 2010;125:369–373. doi: 10.1016/j.jad.2010.01.074. [DOI] [PubMed] [Google Scholar]
- Lee J, Narang P, Enja M, Lippmann S. Use of ketamine in acute cases of suicidality. Innovations in clinical neuroscience. 2015;12:29–31. [PMC free article] [PubMed] [Google Scholar]
- Levenson JM, O’Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. The Journal of biological chemistry. 2004;279:40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- Levenson JM, Roth TL, Lubin FD, Miller CA, Huang IC, Desai P, Malone LM, Sweatt JD. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. The Journal of biological chemistry. 2006;281:15763–15773. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Limon A, Mamdani F, Hjelm BE, Vawter MP, Sequeira A. Targets of polyamine dysregulation in major depression and suicide: Activity-dependent feedback, excitability, and neurotransmission. Neuroscience and biobehavioral reviews. 2016;66:80–91. doi: 10.1016/j.neubiorev.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsky RH, Marini AM. Brain-derived neurotrophic factor in neuronal survival and behavior-related plasticity. Annals of the New York Academy of Sciences. 2007;1122:130–143. doi: 10.1196/annals.1403.009. [DOI] [PubMed] [Google Scholar]
- Lockwood LE, Su S, Youssef NA. The role of epigenetics in depression and suicide: A platform for gene-environment interactions. Psychiatry research. 2015;228:235–242. doi: 10.1016/j.psychres.2015.05.071. [DOI] [PubMed] [Google Scholar]
- Luger K, Dechassa ML, Tremethick DJ. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nature reviews. Molecular cell biology. 2012;13:436–447. doi: 10.1038/nrm3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelli L, Serretti A. Gene-Environment Interaction Studies in Suicidal Behaviour. In: Kaschka WP, RD, editors. Biological Aspects of Suicidal Behavior. Karger; Basel: 2016. pp. 63–74. [Google Scholar]
- Mann JJ. Neurobiology of suicidal behaviour. Nature reviews Neuroscience. 2003;4:819–828. doi: 10.1038/nrn1220. [DOI] [PubMed] [Google Scholar]
- Mann JJ. The serotonergic system in mood disorders and suicidal behaviour. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2013;368:20120537. doi: 10.1098/rstb.2012.0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann JJ, Arango VA, Avenevoli S, Brent DA, Champagne FA, Clayton P, Currier D, Dougherty DM, Haghighi F, Hodge SE, Kleinman J, Lehner T, McMahon F, Moscicki EK, Oquendo MA, Pandey GN, Pearson J, Stanley B, Terwilliger J, Wenzel A. Candidate endophenotypes for genetic studies of suicidal behavior. Biological psychiatry. 2009;65:556–563. doi: 10.1016/j.biopsych.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann JJ, Haghighi F. Genes and environment: multiple pathways to psychopathology. Biological psychiatry. 2010;68:403–404. doi: 10.1016/j.biopsych.2010.07.006. [DOI] [PubMed] [Google Scholar]
- Martinowich K, Lu B. Interaction between BDNF and serotonin: role in mood disorders. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2008;33:73–83. doi: 10.1038/sj.npp.1301571. [DOI] [PubMed] [Google Scholar]
- Martinowich K, Manji H, Lu B. New insights into BDNF function in depression and anxiety. Nature neuroscience. 2007;10:1089–1093. doi: 10.1038/nn1971. [DOI] [PubMed] [Google Scholar]
- Maussion G, Yang J, Suderman M, Diallo A, Nagy C, Arnovitz M, Mechawar N, Turecki G. Functional DNA methylation in a transcript specific 3′UTR region of TrkB associates with suicide. Epigenetics. 2014;9:1061–1070. doi: 10.4161/epi.29068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maussion G, Yang J, Yerko V, Barker P, Mechawar N, Ernst C, Turecki G. Regulation of a truncated form of tropomyosin-related kinase B (TrkB) by Hsa-miR-185* in frontal cortex of suicide completers. PloS one. 2012;7:e39301. doi: 10.1371/journal.pone.0039301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Noh KM, Allis CD. Histone regulation in the CNS: basic principles of epigenetic plasticity. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2013;38:3–22. doi: 10.1038/npp.2012.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGirr A, Alda M, Seguin M, Cabot S, Lesage A, Turecki G. Familial aggregation of suicide explained by cluster B traits: a three-group family study of suicide controlling for major depressive disorder. The American journal of psychiatry. 2009;166:1124–1134. doi: 10.1176/appi.ajp.2009.08111744. [DOI] [PubMed] [Google Scholar]
- McGowan PO, Sasaki A, D’Alessio AC, Dymov S, Labonte B, Szyf M, Turecki G, Meaney MJ. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nature neuroscience. 2009;12:342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan PO, Szyf M. The epigenetics of social adversity in early life: implications for mental health outcomes. Neurobiol Dis. 2010;39:66–72. doi: 10.1016/j.nbd.2009.12.026. [DOI] [PubMed] [Google Scholar]
- McNally L, Bhagwagar Z, Hannestad J. Inflammation, glutamate, and glia in depression: a literature review. CNS spectrums. 2008;13:501–510. doi: 10.1017/s1092852900016734. [DOI] [PubMed] [Google Scholar]
- Mehta D, Gonik M, Klengel T, Rex-Haffner M, Menke A, Rubel J, Mercer KB, Putz B, Bradley B, Holsboer F, Ressler KJ, Muller-Myhsok B, Binder EB. Using polymorphisms in FKBP5 to define biologically distinct subtypes of posttraumatic stress disorder: evidence from endocrine and gene expression studies. Archives of general psychiatry. 2011;68:901–910. doi: 10.1001/archgenpsychiatry.2011.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melhem NM, Keilp JG, Porta G, Oquendo MA, Burke A, Stanley B, Cooper TB, Mann JJ, Brent DA. Blunted HPA Axis Activity in Suicide Attempters Compared to those at High Risk for Suicidal Behavior. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2016;41:1447–1456. doi: 10.1038/npp.2015.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke A, Klengel T, Rubel J, Bruckl T, Pfister H, Lucae S, Uhr M, Holsboer F, Binder EB. Genetic variation in FKBP5 associated with the extent of stress hormone dysregulation in major depression. Genes, brain, and behavior. 2013;12:289–296. doi: 10.1111/gbb.12026. [DOI] [PubMed] [Google Scholar]
- Merali Z, Du L, Hrdina P, Palkovits M, Faludi G, Poulter MO, Anisman H. Dysregulation in the suicide brain: mRNA expression of corticotropin-releasing hormone receptors and GABA(A) receptor subunits in frontal cortical brain region. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2004;24:1478–1485. doi: 10.1523/JNEUROSCI.4734-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minichiello L. TrkB signalling pathways in LTP and learning. Nature reviews Neuroscience. 2009;10:850–860. doi: 10.1038/nrn2738. [DOI] [PubMed] [Google Scholar]
- Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2013;38:23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munkholm K, Peijs L, Kessing LV, Vinberg M. Reduced mRNA expression of PTGDS in peripheral blood mononuclear cells of rapid-cycling bipolar disorder patients compared with healthy control subjects. The international journal of neuropsychopharmacology/official scientific journal of the Collegium Internationale Neuropsychopharmacologicum (CINP) 2015:18. doi: 10.1093/ijnp/pyu101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy C, Suderman M, Yang J, Szyf M, Mechawar N, Ernst C, Turecki G. Astrocytic abnormalities and global DNA methylation patterns in depression and suicide. Molecular psychiatry. 2015;20:320–328. doi: 10.1038/mp.2014.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy C, Torres-Platas SG, Mechawar N, Turecki G. Repression of Astrocytic Connexins in Cortical and Subcortical Brain Regions and Prefrontal Enrichment of H3K9me3 in Depression and Suicide. The international journal of neuropsychopharmacology/official scientific journal of the Collegium Internationale Neuropsychopharmacologicum (CINP) 2016 doi: 10.1093/ijnp/pyw071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan P, Dragunow M. Pharmacology of epigenetics in brain disorders. British journal of pharmacology. 2010;159:285–303. doi: 10.1111/j.1476-5381.2009.00526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebbioso A, Carafa V, Benedetti R, Altucci L. Trials with ‘epigenetic’ drugs: an update. Molecular oncology. 2012;6:657–682. doi: 10.1016/j.molonc.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman EA. Glial cell inhibition of neurons by release of ATP. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2003;23:1659–1666. doi: 10.1523/JNEUROSCI.23-05-01659.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nghia NA, Hirasawa T, Kasai H, Obata C, Moriishi K, Mochizuki K, Koizumi S, Kubota T. Long-term imipramine treatment increases N-methyl-d-aspartate receptor activity and expression via epigenetic mechanisms. European journal of pharmacology. 2015;752:69–77. doi: 10.1016/j.ejphar.2015.02.010. [DOI] [PubMed] [Google Scholar]
- Niciu MJ, Grunschel BD, Corlett PR, Pittenger C, Bloch MH. Two cases of delayed-onset suicidal ideation, dysphoria and anxiety after ketamine infusion in patients with obsessive-compulsive disorder and a history of major depressive disorder. Journal of psychopharmacology (Oxford, England) 2013;27:651–654. doi: 10.1177/0269881113486718. [DOI] [PubMed] [Google Scholar]
- Niculescu AB, Levey D, Le-Niculescu H, Niculescu E, Kurian SM, Salomon D. Psychiatric blood biomarkers: avoiding jumping to premature negative or positive conclusions. Molecular psychiatry. 2015a;20:286–288. doi: 10.1038/mp.2014.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niculescu AB, Levey DF, Phalen PL, Le-Niculescu H, Dainton HD, Jain N, Belanger E, James A, George S, Weber H, Graham DL, Schweitzer R, Ladd TB, Learman R, Niculescu EM, Vanipenta NP, Khan FN, Mullen J, Shankar G, Cook S, Humbert C, Ballew A, Yard M, Gelbart T, Shekhar A, Schork NJ, Kurian SM, Sandusky GE, Salomon DR. Understanding and predicting suicidality using a combined genomic and clinical risk assessment approach. Molecular psychiatry. 2015b;20:1266–1285. doi: 10.1038/mp.2015.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olk S, Zoidl G, Dermietzel R. Connexins, cell motility, and the cytoskeleton. Cell motility and the cytoskeleton. 2009;66:1000–1016. doi: 10.1002/cm.20404. [DOI] [PubMed] [Google Scholar]
- Oquendo MA, Galfalvy HC, Currier D, Grunebaum MF, Sher L, Sullivan GM, Burke AK, Harkavy-Friedman J, Sublette ME, Parsey RV, Mann JJ. Treatment of suicide attempters with bipolar disorder: a randomized clinical trial comparing lithium and valproate in the prevention of suicidal behavior. The American journal of psychiatry. 2011;168:1050–1056. doi: 10.1176/appi.ajp.2011.11010163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paakinaho V, Makkonen H, Jaaskelainen T, Palvimo JJ. Glucocorticoid receptor activates poised FKBP51 locus through long-distance interactions. Molecular endocrinology (Baltimore, Md) 2010;24:511–525. doi: 10.1210/me.2009-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabba M, Sibille E. GABA, Depression and Suicide. In: Kaschka WP, RD, editors. Biological Aspects of Suicidal Behavior. Karger; Basel: 2016. pp. 37–50. [Google Scholar]
- Panatier A, Theodosis DT, Mothet JP, Touquet B, Pollegioni L, Poulain DA, Oliet SH. Glia-derived D-serine controls NMDA receptor activity and synaptic memory. Cell. 2006;125:775–784. doi: 10.1016/j.cell.2006.02.051. [DOI] [PubMed] [Google Scholar]
- Pandey GN. Biological basis of suicide and suicidal behavior. Bipolar Disord. 2013;15:524–541. doi: 10.1111/bdi.12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey GN, Dwivedi Y. What can post-mortem studies tell us about the pathoetiology of suicide? Future neurology. 2010;5:701–720. doi: 10.2217/fnl.10.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey GN, Ren X, Rizavi HS, Conley RR, Roberts RC, Dwivedi Y. Brain-derived neurotrophic factor and tyrosine kinase B receptor signalling in post-mortem brain of teenage suicide victims. The international journal of neuropsychopharmacology/official scientific journal of the Collegium Internationale Neuropsychopharmacologicum (CINP) 2008;11:1047–1061. doi: 10.1017/S1461145708009000. [DOI] [PubMed] [Google Scholar]
- Pandey GN, Rizavi HS, Ren X, Fareed J, Hoppensteadt DA, Roberts RC, Conley RR, Dwivedi Y. Proinflammatory cytokines in the prefrontal cortex of teenage suicide victims. Journal of psychiatric research. 2012;46:57–63. doi: 10.1016/j.jpsychires.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankratov Y, Lalo U. Role for astroglial alpha1-adrenoreceptors in gliotransmission and control of synaptic plasticity in the neocortex. Frontiers in cellular neuroscience. 2015;9:230. doi: 10.3389/fncel.2015.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Poo MM. Neurotrophin regulation of neural circuit development and function. Nature reviews Neuroscience. 2013;14:7–23. doi: 10.1038/nrn3379. [DOI] [PubMed] [Google Scholar]
- Parkhurst CN, Yang G, Ninan I, Savas JN, Yates JR, 3rd, Lafaille JJ, Hempstead BL, Littman DR, Gan WB. Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell. 2013;155:1596–1609. doi: 10.1016/j.cell.2013.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual O, Ben Achour S, Rostaing P, Triller A, Bessis A. Microglia activation triggers astrocyte-mediated modulation of excitatory neurotransmission. Proc Natl Acad Sci U S A. 2012;109:E197–205. doi: 10.1073/pnas.1111098109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual O, Casper KB, Kubera C, Zhang J, Revilla-Sanchez R, Sul JY, Takano H, Moss SJ, McCarthy K, Haydon PG. Astrocytic purinergic signaling coordinates synaptic networks. Science. 2005;310:113–116. doi: 10.1126/science.1116916. [DOI] [PubMed] [Google Scholar]
- Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nature reviews Molecular cell biology. 2013;14:341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson-Leary J, Osborne DM, McNay EC. Role of Glia in Stress-Induced Enhancement and Impairment of Memory. Frontiers in integrative neuroscience. 2015;9:63. doi: 10.3389/fnint.2015.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen NL, Fiske A. Genetic influences on suicide and nonfatal suicidal behavior: twin study findings. European psychiatry: the journal of the Association of European Psychiatrists. 2010;25:264–267. doi: 10.1016/j.eurpsy.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena-Hernandez R, Marques M, Hilmi K, Zhao T, Saad A, Alaoui-Jamali MA, del Rincon SV, Ashworth T, Roy AL, Emerson BM, Witcher M. Genome-wide targeting of the epigenetic regulatory protein CTCF to gene promoters by the transcription factor TFII-I. Proc Natl Acad Sci U S A. 2015;112:E677–686. doi: 10.1073/pnas.1416674112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Ortiz JM, Garcia-Gutierrez MS, Navarrete F, Giner S, Manzanares J. Gene and protein alterations of FKBP5 and glucocorticoid receptor in the amygdala of suicide victims. Psychoneuroendocrinology. 2013;38:1251–1258. doi: 10.1016/j.psyneuen.2012.11.008. [DOI] [PubMed] [Google Scholar]
- Poulter MO, Du L, Weaver IC, Palkovits M, Faludi G, Merali Z, Szyf M, Anisman H. GABAA receptor promoter hypermethylation in suicide brain: implications for the involvement of epigenetic processes. Biological psychiatry. 2008;64:645–652. doi: 10.1016/j.biopsych.2008.05.028. [DOI] [PubMed] [Google Scholar]
- Price RB, Iosifescu DV, Murrough JW, Chang LC, Al Jurdi RK, Iqbal SZ, Soleimani L, Charney DS, Foulkes AL, Mathew SJ. Effects of ketamine on explicit and implicit suicidal cognition: a randomized controlled trial in treatment-resistant depression. Depression and anxiety. 2014;31:335–343. doi: 10.1002/da.22253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price RB, Mathew SJ. Does ketamine have anti-suicidal properties? Current status and future directions. CNS drugs. 2015;29:181–188. doi: 10.1007/s40263-015-0232-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi XR, Kamphuis W, Wang S, Wang Q, Lucassen PJ, Zhou JN, Swaab DF. Aberrant stress hormone receptor balance in the human prefrontal cortex and hypothalamic paraventricular nucleus of depressed patients. Psychoneuroendocrinology. 2013;38:863–870. doi: 10.1016/j.psyneuen.2012.09.014. [DOI] [PubMed] [Google Scholar]
- Raine A, Meloy JR, Bihrle S, Stoddard J, LaCasse L, Buchsbaum MS. Reduced prefrontal and increased subcortical brain functioning assessed using positron emission tomography in predatory and affective murderers. Behavioral sciences & the law. 1998;16:319–332. doi: 10.1002/(sici)1099-0798(199822)16:3<319::aid-bsl311>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Rajkowska G, Stockmeier CA. Astrocyte pathology in major depressive disorder: insights from human postmortem brain tissue. Current drug targets. 2013;14:1225–1236. doi: 10.2174/13894501113149990156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao P, Benito E, Fischer A. MicroRNAs as biomarkers for CNS disease. Frontiers in molecular neuroscience. 2013;6:39. doi: 10.3389/fnmol.2013.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raust A, Slama F, Mathieu F, Roy I, Chenu A, Koncke D, Fouques D, Jollant F, Jouvent E, Courtet P, Leboyer M, Bellivier F. Prefrontal cortex dysfunction in patients with suicidal behavior. Psychological medicine. 2007;37:411–419. doi: 10.1017/S0033291706009111. [DOI] [PubMed] [Google Scholar]
- Rebola N, Simoes AP, Canas PM, Tome AR, Andrade GM, Barry CE, Agostinho PM, Lynch MA, Cunha RA. Adenosine A2A receptors control neuroinflammation and consequent hippocampal neuronal dysfunction. Journal of neurochemistry. 2011;117:100–111. doi: 10.1111/j.1471-4159.2011.07178.x. [DOI] [PubMed] [Google Scholar]
- Reinstatler L, Youssef NA. Ketamine as a potential treatment for suicidal ideation: a systematic review of the literature. Drugs in R&D. 2015;15:37–43. doi: 10.1007/s40268-015-0081-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reus GZ, Abelaira HM, dos Santos MA, Carlessi AS, Tomaz DB, Neotti MV, Liranco JL, Gubert C, Barth M, Kapczinski F, Quevedo J. Ketamine and imipramine in the nucleus accumbens regulate histone deacetylation induced by maternal deprivation and are critical for associated behaviors. Behavioural brain research. 2013;256:451–456. doi: 10.1016/j.bbr.2013.08.041. [DOI] [PubMed] [Google Scholar]
- Rial D, Lemos C, Pinheiro H, Duarte JM, Goncalves FQ, Real JI, Prediger RD, Goncalves N, Gomes CA, Canas PM, Agostinho P, Cunha RA. Depression as a Glial-Based Synaptic Dysfunction. Frontiers in cellular neuroscience. 2015;9:521. doi: 10.3389/fncel.2015.00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice L, Waters CE, Eccles J, Garside H, Sommer P, Kay P, Blackhall FH, Zeef L, Telfer B, Stratford I, Clarke R, Singh D, Stevens A, White A, Ray DW. Identification and functional analysis of SKA2 interaction with the glucocorticoid receptor. The Journal of endocrinology. 2008;198:499–509. doi: 10.1677/JOE-08-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins B, Martin MV, Morgan L, Vawter MP. Analysis of whole genome biomarker expression in blood and brain. American journal of medical genetics Part B, Neuropsychiatric genetics: the official publication of the International Society of Psychiatric Genetics. 2010;153b:919–936. doi: 10.1002/ajmg.b.31062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose CR, Blum R, Pichler B, Lepier A, Kafitz KW, Konnerth A. Truncated TrkB-T1 mediates neurotrophin-evoked calcium signalling in glia cells. Nature. 2003;426:74–78. doi: 10.1038/nature01983. [DOI] [PubMed] [Google Scholar]
- Roy A. Gene–Environment Interaction and Suicidal Behavior. In: Dwivedi Y, editor. The Neurobiological Basis of Suicide. CRC Press/Taylor & Francis; Boca Raton (FL): 2012. [Google Scholar]
- Roy A, Gorodetsky E, Yuan Q, Goldman D, Enoch MA. Interaction of FKBP5, a stress-related gene, with childhood trauma increases the risk for attempting suicide. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2010;35:1674–1683. doi: 10.1038/npp.2009.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Hodgkinson CA, Deluca V, Goldman D, Enoch MA. Two HPA axis genes, CRHBP and FKBP5, interact with childhood trauma to increase the risk for suicidal behavior. Journal of psychiatric research. 2012;46:72–79. doi: 10.1016/j.jpsychires.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Segal NL. Suicidal behavior in twins: a replication. J Affect Disord. 2001;66:71–74. doi: 10.1016/s0165-0327(00)00275-5. [DOI] [PubMed] [Google Scholar]
- Roy A, Segal NL, Centerwall BS, Robinette CD. Suicide in twins. Archives of general psychiatry. 1991;48:29–32. doi: 10.1001/archpsyc.1991.01810250031003. [DOI] [PubMed] [Google Scholar]
- Roy A, Segal NL, Sarchiapone M. Attempted suicide among living co-twins of twin suicide victims. The American journal of psychiatry. 1995;152:1075–1076. doi: 10.1176/ajp.152.7.1075. [DOI] [PubMed] [Google Scholar]
- Sadeh N, Wolf EJ, Logue MW, Hayes JP, Stone A, Griffin LM, Schichman SA, Miller MW. EPIGENETIC VARIATION AT SKA2 PREDICTS SUICIDE PHENOTYPES AND INTERNALIZING PSYCHOPATHOLOGY. Depression and anxiety. 2016;33:308–315. doi: 10.1002/da.22480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterlee JS, Basanta-Sanchez M, Blanco S, Li JB, Meyer K, Pollock J, Sadri-Vakili G, Rybak-Wolf A. Novel RNA modifications in the nervous system: form and function. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2014;34:15170–15177. doi: 10.1523/JNEUROSCI.3236-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scianni M, Antonilli L, Chece G, Cristalli G, Di Castro MA, Limatola C, Maggi L. Fractalkine (CX3CL1) enhances hippocampal N-methyl-D-aspartate receptor (NMDAR) function via D-serine and adenosine receptor type A2 (A2AR) activity. Journal of neuroinflammation. 2013;10:108. doi: 10.1186/1742-2094-10-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira A, Gwadry FG, Ffrench-Mullen JM, Canetti L, Gingras Y, Casero RA, Jr, Rouleau G, Benkelfat C, Turecki G. Implication of SSAT by gene expression and genetic variation in suicide and major depression. Archives of general psychiatry. 2006;63:35–48. doi: 10.1001/archpsyc.63.1.35. [DOI] [PubMed] [Google Scholar]
- Sequeira A, Klempan T, Canetti L, ffrench-Mullen J, Benkelfat C, Rouleau GA, Turecki G. Patterns of gene expression in the limbic system of suicides with and without major depression. Molecular psychiatry. 2007;12:640–655. doi: 10.1038/sj.mp.4001969. [DOI] [PubMed] [Google Scholar]
- Sequeira A, Mamdani F, Ernst C, Vawter MP, Bunney WE, Lebel V, Rehal S, Klempan T, Gratton A, Benkelfat C, Rouleau GA, Mechawar N, Turecki G. Global brain gene expression analysis links glutamatergic and GABAergic alterations to suicide and major depression. PLoS One. 2009;4:e6585. doi: 10.1371/journal.pone.0006585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano A, Haddjeri N, Lacaille JC, Robitaille R. GABAergic network activation of glial cells underlies hippocampal heterosynaptic depression. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2006;26:5370–5382. doi: 10.1523/JNEUROSCI.5255-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma RP. Blood chromatin as a biosensor of the epigenetic milieu: a tool for studies in living psychiatric patients. Epigenomics. 2012;4:551–559. doi: 10.2217/epi.12.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slap G, Goodman E, Huang B. Adoption as a risk factor for attempted suicide during adolescence. Pediatrics. 2001;108:E30. doi: 10.1542/peds.108.2.e30. [DOI] [PubMed] [Google Scholar]
- Smalheiser NR, Lugli G, Rizavi HS, Torvik VI, Turecki G, Dwivedi Y. MicroRNA expression is down-regulated and reorganized in prefrontal cortex of depressed suicide subjects. PLoS One. 2012;7:e33201. doi: 10.1371/journal.pone.0033201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Vale WW. The role of the hypothalamic-pituitary-adrenal axis in neuroendocrine responses to stress. Dialogues in clinical neuroscience. 2006;8:383–395. doi: 10.31887/DCNS.2006.8.4/ssmith. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speckens AE, Hawton K. Social problem solving in adolescents with suicidal behavior: a systematic review. Suicide & life-threatening behavior. 2005;35:365–387. doi: 10.1521/suli.2005.35.4.365. [DOI] [PubMed] [Google Scholar]
- Sun H, Kennedy PJ, Nestler EJ. Epigenetics of the depressed brain: role of histone acetylation and methylation. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2013;38:124–137. doi: 10.1038/npp.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JD, Liu Y, Yuan YH, Li J, Chen NH. Gap junction dysfunction in the prefrontal cortex induces depressive-like behaviors in rats. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2012;37:1305–1320. doi: 10.1038/npp.2011.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Zang L, Shu Q, Li X. From development to diseases: the role of 5hmC in brain. Genomics. 2014;104:347–351. doi: 10.1016/j.ygeno.2014.08.021. [DOI] [PubMed] [Google Scholar]
- Supriyanto I, Sasada T, Fukutake M, Asano M, Ueno Y, Nagasaki Y, Shirakawa O, Hishimoto A. Association of FKBP5 gene haplotypes with completed suicide in the Japanese population. Progress in neuro-psychopharmacology & biological psychiatry. 2011;35:252–256. doi: 10.1016/j.pnpbp.2010.11.019. [DOI] [PubMed] [Google Scholar]
- Szyf M. Epigenetics, DNA methylation, and chromatin modifying drugs. Annual review of pharmacology and toxicology. 2009;49:243–263. doi: 10.1146/annurev-pharmtox-061008-103102. [DOI] [PubMed] [Google Scholar]
- Tan L, Shi YG. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development (Cambridge, England) 2012;139:1895–1902. doi: 10.1242/dev.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatro ET, Everall IP, Kaul M, Achim CL. Modulation of glucocorticoid receptor nuclear translocation in neurons by immunophilins FKBP51 and FKBP52: implications for major depressive disorder. Brain research. 2009;1286:1–12. doi: 10.1016/j.brainres.2009.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakurta RG, Das R, Bhattacharya AK, Saha D, Sen S, Singh OP, Bisui B. Rapid response with ketamine on suicidal cognition in resistant depression. Indian journal of psychological medicine. 2012;34:170–175. doi: 10.4103/0253-7176.101793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Platas SG, Hercher C, Davoli MA, Maussion G, Labonte B, Turecki G, Mechawar N. Astrocytic hypertrophy in anterior cingulate white matter of depressed suicides. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2011;36:2650–2658. doi: 10.1038/npp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripp A, Oh H, Guilloux JP, Martinowich K, Lewis DA, Sibille E. Brain-derived neurotrophic factor signaling and subgenual anterior cingulate cortex dysfunction in major depressive disorder. The American journal of psychiatry. 2012;169:1194–1202. doi: 10.1176/appi.ajp.2012.12020248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankova N, Renthal W, Kumar A, Nestler EJ. Epigenetic regulation in psychiatric disorders. Nature reviews Neuroscience. 2007;8:355–367. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, Nestler EJ. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nature neuroscience. 2006;9:519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- Turecki G. The molecular bases of the suicidal brain. Nature reviews Neuroscience. 2014;15:802–816. doi: 10.1038/nrn3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heeringen C, Bijttebier S, Godfrin K. Suicidal brains: a review of functional and structural brain studies in association with suicidal behaviour. Neuroscience and biobehavioral reviews. 2011;35:688–698. doi: 10.1016/j.neubiorev.2010.08.007. [DOI] [PubMed] [Google Scholar]
- Van Heeringen C, Marusic A. Understanding the suicidal brain. The British journal of psychiatry: the journal of mental science. 2003;183:282–284. doi: 10.1192/bjp.183.4.282. [DOI] [PubMed] [Google Scholar]
- van Heeringen K. The neurobiology of suicide and suicidality. Canadian journal of psychiatry Revue canadienne de psychiatrie. 2003;48:292–300. doi: 10.1177/070674370304800504. [DOI] [PubMed] [Google Scholar]
- Venkatesh S, Workman JL. Histone exchange, chromatin structure and the regulation of transcription. Nature reviews Molecular cell biology. 2015;16:178–189. doi: 10.1038/nrm3941. [DOI] [PubMed] [Google Scholar]
- Vermeer H, Hendriks-Stegeman BI, van der Burg B, van Buul-Offers SC, Jansen M. Glucocorticoid-induced increase in lymphocytic FKBP51 messenger ribonucleic acid expression: a potential marker for glucocorticoid sensitivity, potency, and bioavailability. The Journal of clinical endocrinology and metabolism. 2003;88:277–284. doi: 10.1210/jc.2002-020354. [DOI] [PubMed] [Google Scholar]
- Vialou V, Feng J, Robison AJ, Nestler EJ. Epigenetic mechanisms of depression and antidepressant action. Annual review of pharmacology and toxicology. 2013;53:59–87. doi: 10.1146/annurev-pharmtox-010611-134540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nature neuroscience. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Weaver IC, D’Alessio AC, Brown SE, Hellstrom IC, Dymov S, Sharma S, Szyf M, Meaney MJ. The transcription factor nerve growth factor-inducible protein a mediates epigenetic programming: altering epigenetic marks by immediate-early genes. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2007;27:1756–1768. doi: 10.1523/JNEUROSCI.4164-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Preventing suicide: A global imperative. 2014. [Google Scholar]
- Wilkinson MB, Xiao G, Kumar A, LaPlant Q, Renthal W, Sikder D, Kodadek TJ, Nestler EJ. Imipramine treatment and resiliency exhibit similar chromatin regulation in the mouse nucleus accumbens in depression models. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2009;29:7820–7832. doi: 10.1523/JNEUROSCI.0932-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson ST, Sanacora G. KETAMINE: A POTENTIAL RAPID-ACTING ANTISUICIDAL AGENT? Depression and anxiety. 2016;33:711–717. doi: 10.1002/da.22498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willour VL, Chen H, Toolan J, Belmonte P, Cutler DJ, Goes FS, Zandi PP, Lee RS, MacKinnon DF, Mondimore FM, Schweizer B, DePaulo JR, Jr, Gershon ES, McMahon FJ, Potash JB. Family-based association of FKBP5 in bipolar disorder. Molecular psychiatry. 2009;14:261–268. doi: 10.1038/sj.mp.4002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Chen PS, Dallas S, Wilson B, Block ML, Wang CC, Kinyamu H, Lu N, Gao X, Leng Y, Chuang DM, Zhang W, Lu RB, Hong JS. Histone deacetylase inhibitors up-regulate astrocyte GDNF and BDNF gene transcription and protect dopaminergic neurons. The international journal of neuropsychopharmacology/official scientific journal of the Collegium Internationale Neuropsychopharmacologicum (CINP) 2008;11:1123–1134. doi: 10.1017/S1461145708009024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Ge W, Chen Y, Zhang Z, Shen W, Wu C, Poo M, Duan S. Contribution of astrocytes to hippocampal long-term potentiation through release of D-serine. Proc Natl Acad Sci U S A. 2003;100:15194–15199. doi: 10.1073/pnas.2431073100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao I, Iida J, Nishimura W, Hata Y. Synaptic localization of SAPAP1, a synaptic membrane-associated protein. Genes to cells: devoted to molecular & cellular mechanisms. 2003;8:121–129. doi: 10.1046/j.1365-2443.2003.00622.x. [DOI] [PubMed] [Google Scholar]
- Yu H, Su Y, Shin J, Zhong C, Guo JU, Weng YL, Gao F, Geschwind DH, Coppola G, Ming GL, Song H. Tet3 regulates synaptic transmission and homeostatic plasticity via DNA oxidation and repair. Nature neuroscience. 2015;18:836–843. doi: 10.1038/nn.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zai CC. Genetic Factors and Suicidal Behavior. In: Dwivedi Y, editor. The Neurobiological Basis of Suicide. Frontiers in Neuroscience. CRC Press/Taylor & Francis; Boca Raton (FL): 2012. [Google Scholar]
- Zannas AS, Binder EB. Gene-environment interactions at the FKBP5 locus: sensitive periods, mechanisms and pleiotropism. Genes, brain, and behavior. 2014;13:25–37. doi: 10.1111/gbb.12104. [DOI] [PubMed] [Google Scholar]
- Zannas AS, Wiechmann T, Gassen NC, Binder EB. Gene-Stress-Epigenetic Regulation of FKBP5: Clinical and Translational Implications. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2016;41:261–274. doi: 10.1038/npp.2015.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan Y, Paolicelli RC, Sforazzini F, Weinhard L, Bolasco G, Pagani F, Vyssotski AL, Bifone A, Gozzi A, Ragozzino D, Gross CT. Deficient neuron-microglia signaling results in impaired functional brain connectivity and social behavior. Nature neuroscience. 2014;17:400–406. doi: 10.1038/nn.3641. [DOI] [PubMed] [Google Scholar]
- Zhang JM, Wang HK, Ye CQ, Ge W, Chen Y, Jiang ZL, Wu CP, Poo MM, Duan S. ATP released by astrocytes mediates glutamatergic activity-dependent heterosynaptic suppression. Neuron. 2003;40:971–982. doi: 10.1016/s0896-6273(03)00717-7. [DOI] [PubMed] [Google Scholar]
- Zheleznyakova GY, Nilsson EK, Kiselev AV, Maretina MA, Tishchenko LI, Fredriksson R, Baranov VS, Schioth HB. Methylation levels of SLC23A2 and NCOR2 genes correlate with spinal muscular atrophy severity. PloS one. 2015;10:e0121964. doi: 10.1371/journal.pone.0121964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Bruckl T, Nocon A, Pfister H, Binder EB, Uhr M, Lieb R, Moffitt TE, Caspi A, Holsboer F, Ising M. Interaction of FKBP5 gene variants and adverse life events in predicting depression onset: results from a 10-year prospective community study. The American journal of psychiatry. 2011;168:1107–1116. doi: 10.1176/appi.ajp.2011.10111577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Tan Y, Wang Z, Wang KS, Zhang X, Chen X, Li CS, Wang T, Luo X. Long noncoding RNAs in psychiatric disorders. Psychiatric genetics. 2016;26:109–116. doi: 10.1097/YPG.0000000000000129. [DOI] [PMC free article] [PubMed] [Google Scholar]