Abstract
Campylobacter jejuni is the leading cause of bacterial foodborne diarrhoeal disease worldwide. Despite the microaerophilic nature of the bacterium, C. jejuni can survive the atmospheric oxygen conditions in the environment. Bacteria that can survive either within a host or in the environment like C. jejuni require variable responses to survive the stresses associated with exposure to different levels of reactive oxygen species. The MarR-type transcriptional regulators RrpA and RrpB have recently been shown to play a role in controlling both the C. jejuni oxidative and aerobic stress responses. Analysis of 3,746 C. jejuni and 486 C. coli genome sequences showed that whilst rrpA is present in over 99% of C. jejuni strains, the presence of rrpB is restricted and appears to correlate with specific MLST clonal complexes (predominantly ST-21 and ST-61). C. coli strains in contrast lack both rrpA and rrpB. In C. jejuni rrpB+ strains, the rrpB gene is located within a variable genomic region containing the IF subtype of the type I Restriction-Modification (hsd) system, whilst this variable genomic region in C. jejuni rrpB- strains contains the IAB subtype hsd system and not the rrpB gene. C. jejuni rrpB- strains exhibit greater resistance to peroxide and aerobic stress than C. jejuni rrpB+ strains. Inactivation of rrpA resulted in increased sensitivity to peroxide stress in rrpB+ strains, but not in rrpB- strains. Mutation of rrpA resulted in reduced killing of Galleria mellonella larvae and enhanced biofilm formation independent of rrpB status. The oxidative and aerobic stress responses of rrpB- and rrpB+ strains suggest adaptation of C. jejuni within different hosts and niches that can be linked to specific MLST clonal complexes.
Keywords: Campylobacter jejuni oxidative stress response regulation, aerobic stress response regulation, transcription factors, restriction modification system
Introduction
Campylobacter jejuni is the leading cause of bacterial foodborne diarrhoeal disease worldwide with an estimated 400 million human infections occurring each year (Ruiz-Palacios, 2007). The predominance of C. jejuni can be attributed to the ability to survive in the environment as well as within avian and mammalian hosts despite the microaerophilic nature of this bacterium (Byrne et al., 2007). C. jejuni has evolved specific adaptation mechanisms to survive under atmospheric oxygen conditions (Kim et al., 2015). In addition to aerobic stress such as the exposure to increased levels of oxygen under atmospheric conditions, C. jejuni can also encounter stress conditions within the host, specifically oxidative stress in the form of reactive oxygen species (ROS) during in vivo survival (Fang, 2004; Palyada et al., 2009). ROS is a collective term that describes the chemical species generated upon incomplete reduction of oxygen (Imlay, 2003) with examples including the superoxide anion (O2-), hydrogen peroxide (H2O2) and the hydroxyl radical (•OH) (D’Autreaux and Toledano, 2007). The accumulation of ROS in the bacterial cytoplasm and periplasm leads to damage of nucleic acids, proteins and membrane structures (Atack and Kelly, 2009).
Bacteria that can survive either within a host or in the environment like C. jejuni require variable responses to survive the stresses associated with exposure to different levels of ROS (Kim et al., 2015). Therefore it is not surprising that C. jejuni contains a number of regulatory proteins involved in the oxidative stress response such as PerR (Handley et al., 2015), Fur (van Vliet et al., 2000), and CosR (Hwang et al., 2011). The C. jejuni NCTC 11168 genome also contains two MarR-type transcriptional regulators that have previously been designated as RrpA and RrpB (Gundogdu et al., 2011, 2015). Using C. jejuni 11168H [a hypermotile derivative of the original sequenced strain NCTC 11168 that shows higher levels of cecal colonization in a chick colonization model (Karlyshev et al., 2002; Jones et al., 2004)] we have shown that both RrpA and RrpB play a role in oxidative and aerobic stress responses with auto-regulatory activity typical of MarR-type transcriptional regulators (Gundogdu et al., 2011, 2015). In addition, RrpA has also been shown to bind upstream of katA suggesting that RrpA directly influences the expression of catalase (KatA). Both 11168H rrpA and rrpB mutants exhibited reduced KatA activity. However, a 11168H rrpAB double mutant exhibited higher levels of resistance to hydrogen peroxide oxidative stress, but similar levels of KatA activity compared to the wild-type strain. Neither the 11168H rrpA mutant nor the 11168H rrpB mutant exhibited any significant difference in sensitivity to either cumene hydroperoxide or menadione oxidative stresses, but both mutants exhibited reduced cytotoxicity in the Galleria mellonella model of infection and enhanced biofilm formation. However, the 11168H rrpAB double mutant exhibited wild-type levels of both cytotoxicity in the G. mellonella model of infection and biofilm formation. Together these data indicate a role for both RrpA and RrpB in the C. jejuni oxidative and aerobic stress responses, enhancing bacterial survival both within a host and in the environment, but also prompted further investigations in order to understand the specific roles of RrpA and RrpB.
Traditional typing methods have failed to identify C. jejuni strains from different sources that cause disease in humans (Champion et al., 2005). Human infections are largely attributed to undercooking of poultry products or poor food hygiene practices involving handling of such produce (Sheppard et al., 2009). C. jejuni can survive in many different niches with the organism isolated from avian, animal, human, and environmental sources (Young et al., 2007). However, whole genome phylogenetic analysis of C. jejuni strains using microarrays has identified different clades and subclades linked to the source of the isolate (Champion et al., 2005; Stabler et al., 2013). Previously we identified differences in the distribution of rrpA and rrpB regulators amongst 111 C. jejuni strains with rrpA present in over 95% and rrpB in approximately only 50% of these strains (Gundogdu et al., 2011). A more recent analysis of 270 C. jejuni strains identified nine clusters (C1–C9) based on genotype and multilocus sequence typing (MLST) data and indicated that clusters C1–C6 were dominated by livestock-associated clonal complexes, whilst clusters C7–C9 contained the majority of the water and wildlife associated clonal complexes (Stabler et al., 2013). Using the original data from this study, rrpA was identified in 129/133 (96.99%) strains within the C1–C6 subclades and in 130/137 (94.89%) strains within the C7–C9 subclades. In contrast rrpB was identified in 102/133 (76.67%) strains within the C1–C6 subclades and only in 19/137 (13.87%) strains within the C7–C9 subclades. In total rrpA was identified in 259/270 (95.92%) strains whilst rrpB was identified in only in 121/270 (44.81%) strains (Supplementary Figure 1; Supplementary Table 1).
The discovery of the varied distribution for rrpA and rrpB has led us to further explore the potential reasons as to why certain strains have one or both regulators. To try and understand the reasons for this, we have investigated the presence or absence of rrpA and rrpB in 4,232 C. jejuni and C. coli genome sequences. Analysis of 4,232 Campylobacter genomes showed that whilst rrpA is present in over 99% of C. jejuni strains, the presence of rrpB is restricted and appears to correlate with livestock-associated MLST clonal complexes. Further analysis showed that the presence of rrpB is linked to a hypervariable region containing the IF subtype of the type I Restriction-Modification (hsd) system, whereas rrpB- strains contain the IAB subtype hsd system. Further investigation of the phenotypes of different C. jejuni rrpB- and rrpB+ strains identified a link between the presence of the MarR-type transcriptional regulator RrpB with the ability of C. jejuni to adapt and survive in different environmental niches. The oxidative and aerobic stress response of rrpB- and rrpB+ strains suggests adaptation of C. jejuni within different hosts and niches that can be linked with specific MLST clonal complexes.
Materials and Methods
Comparative Genomics of rrpA and rrpB within Campylobacter Genomes
A total of 4,232 complete and draft Campylobacter genome sequences (3,746 C. jejuni and 486 C. coli) were obtained from public collections (Jolley and Maiden, 2010; Cody et al., 2013) and are listed in Supplementary Table 2 with accession numbers/pubMLST IDs and assembly status. These genomes were previously used for identification of DNase-genes (Brown et al., 2015), CRISPR repeats and cas genes (Pearson et al., 2015) and the fucose utilization operon (Dwivedi et al., 2016). Genomes included were between 1.5 and 2.0 Mbp and had at least 5 of the 7 MLST alleles identifiable using BLAST. The genomes were phylogenetically clustered using FFPry feature frequency profiling with a word length of 18 (Van Vliet and Kusters, 2015) and further analyzed by MLST (Pearson et al., 2015). Genomes were provisionally annotated using Prokka (Seemann, 2014) and searched for the presence of the predicted RrpA and RrpB proteins using BLASTP (Supplementary Table 2). Presence of the corresponding genes was also assessed at the DNA level using the MIST program (Kruczkiewicz et al., 2013) and the BLAST+ (v2.28) suite. Both DNA and amino acid comparisons were done using the rrpA genes/RrpA proteins from C. jejuni NCTC 11168, C. jejuni 81116, C. jejuni 414, and C. coli 76639, and the rrpB genes/RrpB proteins of C. jejuni NCTC 11168 and C. coli 2544. Conservation of flanking genes was assessed using the output from the comparative genomics software package Roary (Page et al., 2015) and the provisional Prokka annotation of the 4,232 C. jejuni and C. coli genome sequences (Supplementary Tables 2 and 3).
Bacterial Strains and Growth Conditions
Campylobacter jejuni strains (Table 1) were grown at 37°C in a microaerobic chamber (Don Whitley Scientific, United Kingdom) containing 85% N2, 10% CO2 and 5% O2 either on blood agar (BA) plates containing Columbia agar base (Oxoid, United Kingdom) supplemented with 7% (v/v) horse blood (TCS Microbiology, United Kingdom) and Campylobacter Selective Supplement (Oxoid) or in Brucella broth (Oxoid) with shaking at 75 rpm. C. jejuni strains were grown on BA plates for 24 h prior to use in all assays unless otherwise stated. Escherichia coli XL-2 Blue MRF’ competent cells (Stratagene, United Kingdom) were used for cloning experiments and were grown at 37°C in aerobic conditions either on Luria-Bertani (LB) agar plates (Oxoid) or in LB broth (Oxoid) with shaking at 200 rpm. Antibiotics were added at the following concentrations; ampicillin (100 μg/ml), kanamycin (50 μg/ml), and chloramphenicol (50 μg/ml for E. coli studies or 10 μg/ml for C. jejuni studies). All reagents were obtained from Fisher Scientific (United Kingdom) unless otherwise stated.
Table 1.
Campylobacter jejuni strains used in this study.
| Strain | Description | Reference |
|---|---|---|
| 11168H | A hypermotile derivative of the original sequence strain NCTC 11168 that shows higher levels of caecal colonization in a chick colonization model. C3 clade. MLST clonal complex ST-21. | Karlyshev et al., 2002; Jones et al., 2004 |
| 81-176 | Highly virulent and widely studied laboratory strain of C. jejuni. MLST clonal complex ST-42. | Korlath et al., 1985 |
| 81116 | Genetically stable strain which remains infective in avian models. C9ii clade. MLST clonal complex ST-283. | Wassenaar et al., 1991 |
| M1 | M1 (laboratory designation 99/308) is a rarely documented case of direct transmission of C. jejuni from chicken to a person, resulting in enteritis. C9ii clade. MLST clonal complex ST-45. | Friis et al., 2010 |
| 11168H rrpB mutant | Isogenic 11168H rrpB mutant with insertion of a 1.4 kb KanR cassette | Gundogdu et al., 2011 |
| 11168H rrpB complement | rrpB complement constructed by the insertion of the rrpB CDS into the Cj0233 pseudogene in the 11168H rrpB mutant (pDENNIS complementation vector used) | Gundogdu et al., 2011 |
| 81-176 rrpB mutant | Isogenic 81-176 rrpB mutant with insertion of a 1.4 kb KanR cassette | This study |
| 81-176 rrpB complement | rrpB complement constructed by the insertion of the rrpB CDS into a rRNA gene in the 81-176 rrpB mutant (pRRC complementation vector used) | This study |
| 11168H rrpA mutant | Isogenic 11168H rrpA mutant with insertion of a 1.4 kb KanR cassette. | Gundogdu et al., 2015 |
| 11168H rrpA complement | rrpA complement constructed by the insertion of the rrpA CDS into a rRNA gene in the 11168H rrpA mutant (pRRC complementation vector used). | Gundogdu et al., 2015 |
| 81-176 rrpA mutant | Isogenic 81-176 Cj1546 mutant with insertion of a 1.4 kb KanR cassette | This study |
| 81-176 rrpA complement | rrpA complement constructed by the insertion of the rrpA CDS into a rRNA gene in the 81-176 rrpA mutant (pRRC complementation vector used) | This study |
| 81116 rrpA mutant | Isogenic 81116 rrpA mutant with insertion of a 1.4 kb KanR cassette | This study |
| 81116 rrpA complement | rrpA complement constructed by the insertion of the rrpA CDS into a rRNA gene in the 81116 rrpA mutant (pRRC complementation vector used) | This study |
| M1 rrpA mutant | Isogenic M1 rrpA mutant with insertion of a 1.4 kb KanR cassette | This study |
| 40917 | Clinical bloody diarrhea isolate HS type 21 Water and wildlife associated strain. C9ii clade. MLST clonal complex ST-45. | Campylobacter Reference Lab, UK |
| 12241 | Ovine isolate HS type 50 Water and wildlife associated strain. C9ii clade. MLST clonal complex ST-206. | Campylobacter Reference Lab, UK |
| 64555 | Clinical bloody diarrhea isolate HS type 31 Water and wildlife associated strain. C8 clade. | Campylobacter Reference Lab, UK |
| 47693 | Isolated from chicken isolate HS type 27 Water and wildlife associated strain. C9ii clade. MLST clonal complex ST-45. | Campylobacter Reference Lab, UK |
| 62914 | Clinical vomiting isolate HS type – untypeable Water and wildlife associated strain. C7 clade. | Campylobacter Reference Lab, UK |
| 44119 | Clinical septicaemia isolate HS type – 18 Water and wildlife associated strain. C7 clade. | Campylobacter Reference Lab, UK |
| 32787 | Clinical asymptomatic isolate HS type – 18 Water and wildlife associated strain. C9i clade. | Campylobacter Reference Lab, UK |
| Hi80614 | Human isolate Water and wildlife associated strain. C9i clade. | Campylobacter Reference Lab, UK |
| Hi41100305 | Human isolate Water and wildlife associated strain. C9i clade. | Campylobacter Reference Lab, UK |
| 31481 | Clinical asymptomatic isolate HS type – 37 Water and wildlife associated strain. C8 clade. | Campylobacter Reference Lab, UK |
| 47886 | Clinical septicaemia isolate HS type – untypeable Livestock associated strain. C1 clade. | Campylobacter Reference Lab, UK |
| 30280 | Clinical diarrhea isolate HS type – 16 Livestock associated strain. C2 clade. | Campylobacter Reference Lab, UK |
| 13713 | Ox liver portion isolate HS type – 2 Livestock associated strain. C1 clade. | Campylobacter Reference Lab, UK |
| 11973 | Chicken isolate HS type – 2 Livestock associated strain. C1 clade. | Campylobacter Reference Lab, UK |
| G1 | Clinical GBS isolate HS type – 1 Livestock associated strain. C5 clade. MLST clonal complex ST-21. | Guy’s Hospital, UK |
| Bovine27 | Bovine isolate Livestock associated strain. C2 clade. | University of Bristol, UK |
| 12912 | Ox liver portion isolate HS type – 50 Livestock associated strain. C4 clade. | Campylobacter Reference Lab, UK |
| 13040 | Chicken isolate HS type – 50 Livestock associated strain. C6 clade. | Campylobacter Reference Lab, UK |
| 40209 | Chicken isolate HS type – 5 Livestock associated strain. C4 clade. | Campylobacter Reference Lab, UK |
| 91B1 | Chicken isolate Livestock associated strain. C5 clade. | Oxford University, UK |
| Hi41380304 | Human isolate Livestock associated strain. C3 clade. | Campylobacter Reference Lab, UK |
| 11168H perR mutant | Obtained from Campylobacter mutant bank http://crf.lshtm.ac.uk/wren\_mutants.htm | LSHTM mutant bank |
| 11168H sodB mutant | Obtained from Campylobacter mutant bank http://crf.lshtm.ac.uk/wren\_mutants.htm | LSHTM mutant bank |
| 11168H ahpC mutant | Obtained from Campylobacter mutant bank http://crf.lshtm.ac.uk/wren\_mutants.htm | LSHTM mutant bank |
| 11168H katA mutant | Obtained from Campylobacter mutant bank http://crf.lshtm.ac.uk/wren\_mutants.htm | LSHTM mutant bank |
Construction of C. jejuni Mutants
Campylobacter jejuni mutants were constructed as described previously (Gundogdu et al., 2011). Briefly, genes or gene fragments were amplified from C. jejuni genomic DNA using the appropriate gene specific primers (Table 2). PCR products were ligated with pGEM-T Easy vector (Promega, United Kingdom) and then transformed into XL-2 Blue MRF’ cells. If required, inverse PCR mutagenesis (IPCRM) was performed to introduce a unique BglII site into the cloned gene. A kanamycin cassette (KanR) was then ligated into the unique BglII site within the cloned gene (Trieu-Cuot et al., 1985; van Vliet et al., 1998). These constructs were electroporated into competent C. jejuni cells and putative clones were confirmed by PCR and sequencing as described previously (Gundogdu et al., 2011).
Table 2.
Oligonucleotide primers used in this study.
| Primer Name | Sequence |
|---|---|
| 11168H and 81-176 Cj1556-F | ATCATTCTCTTTGTCCTAT |
| 11168H and 81-176 Cj1556-R | TAAGATGGATTCTAAACTATTG |
| 11168H Comp-Cj1556-F | CCCCCATGGATAAGGATTTATAATGAAAAAATATCATTCTCT |
| 11168H Comp-Cj1556-R | CCCGCTAGCTTAAACGATATTTTTATAGCTAT |
| 81-176 Comp-Cj1556-F | CCCTCTAGAATAAGGATTTATAATGAAAAAATATCATTCTCT |
| 81-176 Comp-Cj1556-R | CCCTCTAGATTAAACGATATTTTTATAGCTAT |
| 11168H and 81-176 Cj1546-F | TACTAGGATTTTCATGAG |
| 11168H and 81-176 Cj1546-R | AGATGTTAAATCTCACTGCT |
| 11168H and 81-176 Cj1546 IPCR-F | GGGAGATCTCTCTTAAGGTATTGGTTA |
| 11168H and 81-176 Cj1546 IPCR-R | GGGAGATCTCGATGGTTTAATTATCAG |
| 11168H and 81-176 Comp-Cj1546-F | CCCTCTAGACTAAAGGAATGTTAAATGACTAAAGAGAATTCTCCG |
| 11168H and 81-176 Comp-Cj1546-R | CCCTCTAGATTAATTCAAGCATTTTTTCCC |
| 11168H and 81-176 Cj1546-F checking primers | TTCTCCGTGCAATTTCG |
| 11168H and 81-176 Cj1546-F checking primers | CCATTTGCTCATAGCTTGTAA |
| 81116 Cj1546-F | ATGAGTTTGATCTACTTCG |
| 81116 Cj1546-R | AGATATTAAATCTCACTGCT |
| 81116 Cj1546 IPCR-F | GGGAGATCTCTCTTAAGGTATTGGTTA |
| 81116 Cj1546 IPCR-R | GGGAGATCTTGATGGTTTAATTATCAG |
| 81116 Comp-Cj1546-F | CCCTCTAGACTAAAGGAATGTTAAATGACTAAAGAGAATTCTCAG |
| 81116 Comp-Cj1546-R | CCCTCTAGATTATCAAGCATTTTTTTCC |
| 81116 Cj1546-F checking primers | ATGACTAAAGAGAATTCTC |
| 81116 Cj1546-R checking primers | CCATTTGCACATAGCTTG |
| KanR forward-out | TGGGTTTCAAGCATTAGTCCATGCAAG |
| KanR reverse-out | GTGGTATGACATTGCCTTCTGCG |
| CatR forward-out | CGATTGATGATCGTTGTA |
| CatR reverse-out | TACAGCAGACTATACTG |
Oxidative Stress and Aerobic Growth Assays
Oxidative stress and aerobic growth assays were performed as described previously (Gundogdu et al., 2011, 2015). Briefly, bacterial cells were harvested into 1 ml PBS and diluted to an OD600 of 1. For oxidative stress assays, bacterial cells were exposed to H2O2 at final concentrations of 25, 50, or 100 mM for 15 min, menadione at a final concentration of 100 mM for 60 min and cumene hydroperoxide at 0.05% (w/v) for 15 min, all at 37°C under microaerobic conditions. Serial dilutions were prepared and 10 μl of the 10-1–10-6 dilutions spotted onto BA plates, incubated for 48 h and colonies counted. For growth curves, 10 ml Brucella broth was pre-incubated in a 30 ml flask at 37°C under microaerobic conditions for 24 h. Bacterial cells grown on BA plates for 24 h were used to inoculate pre-incubated Brucella broth at an OD600 of 0.1 and grown for up to 24 h at 37°C under microaerobic and aerobic conditions. OD600 readings were performed at selected time points. In addition bacterial colony forming units (CFUs) were assessed at time point 16 h under microaerobic and aerobic conditions.
Galleria mellonella Infection Model
Galleria mellonella infection assays were performed as described previously (Champion et al., 2010; Gundogdu et al., 2015). Briefly, G. mellonella larvae (LiveFoods Direct, United Kingdom) were stored at 16°C on wood chips. Ten larvae for each experiment were infected with a 10 μl inoculum of a 24 h C. jejuni culture diluted to OD600 0.1 by micro-injection (Hamilton, Switzerland) in the right foremost leg, giving an infectious dose of approximately 106 CFU (Champion et al., 2010). Controls were injection with PBS and no injection. Larvae were incubated at 37°C with survival recorded at 24 h.
Biofilm Assays
Biofilm assays were performed as described previously (Gundogdu et al., 2015). Briefly, bacterial cells were harvested into Mueller Hinton broth, then inoculated to an OD600 of 0.1 into 10 ml Mueller Hinton broth pre-incubated in a 25 ml flask at 37°C under microaerobic conditions for 24 h prior to inoculation, then grown for 5 h at 37°C under microaerobic conditions with shaking at 75 rpm. The OD600 was readjusted to 0.1, then 1 ml of culture was added to a 24 well polystyrene plate (Corning, U.S.A) and incubated at 37°C under either aerobic or microaerobic conditions stationary for 72 h. The wells were washed twice with PBS, dried for 20 min at 37°C followed by addition of 1% (w/v) crystal violet (Sigma–Aldrich) for 15 min. The wells were washed three times with PBS, then destained with 10% (v/v) acetic acid / 30% (v/v) methanol. Absorbance (A595) was measured using a SpectraMax M3 microplate reader (Molecular Devices, USA).
Statistical Analyses
The data is presented as mean + SD. All experiments were performed with at least three biological replicates. Each biological replicate was performed in three technical replicates. Statistical analyses were performed using Prism software (GraphPad Software). All statistical analyses were performed comparing two data sets directly assuming a normal distribution using a two-way student’s t-test (∗ = p < 0.05, ∗∗ = p < 0.01, ∗∗∗ = p < 0.001).
Results
Presence of rrpA and rrpB in a Collection of 4,232 C. jejuni and C. coli
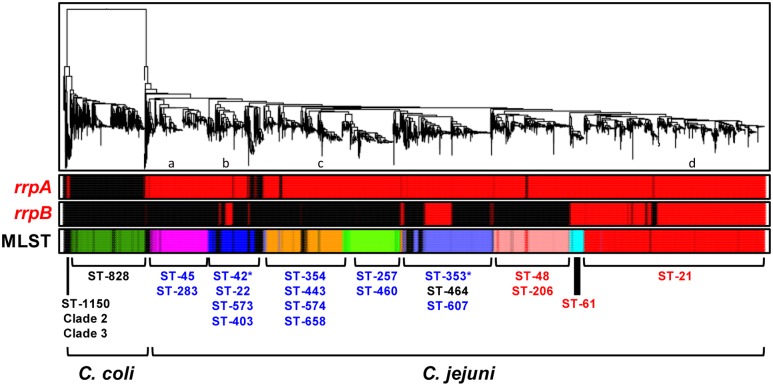
To assess the distribution of rrpA and rrpB in C. jejuni and C. coli we utilized a collection of 4,232 Campylobacter genome sequences (3,746 C. jejuni and 486 C. coli) from public databases (Cody et al., 2013; Brown et al., 2015), which were phylogenetically clustered using FFPry feature frequency profiling (Van Vliet and Kusters, 2015; Dwivedi et al., 2016) and further analyzed for MLST sequence type and clonal complex (Pearson et al., 2015). The vast majority of C. jejuni strains contain rrpA whilst the presence of rrpB is more restricted (Figure 1; Supplementary Table 2 – Sheet 2). The presence or absence of rrpB appears to correlate with MLST clonal complex, especially ST-21 and ST-61. Most C. coli strains contain neither rrpA nor rrpB, with only 12 C. coli genomes encoding an RrpA ortholog and only a single C. coli genome encoding an RrpB ortholog (Supplementary Table 2). Variation in RrpA and RrpB was observed with isolates from C. jejuni MSLT clonal complex ST-607 having a shorter RrpA protein lacking the N-terminal 27 amino acids, while a proportion of C. jejuni ST-353 isolates have a RrpB protein either lacking the N-terminal 29 amino acids, the C-terminal 29 amino acids or a combination of 29 N-terminal amino acids and 9 C-terminal amino acids (Supplementary Figure 2). There was no apparent link between the truncated versions and source of the isolates (Supplementary Table 2), suggesting these are different forms of RrpA and RrpB circulating within clonal complexes.
FIGURE 1.
Prevalence of the rrpA and rrpB genes among 3,746 Campylobacter jejuni and 486 C. coli genomes. Genomes were phylogenetically clustered using FFPry feature frequency profiling with L = 18 (Van Vliet and Kusters, 2015). The first two rows labeled ‘rrpA’ and ‘rrpB’ indicate genomes possessing the respective genes, which are shown in red, while those lacking the pathway are shown in black. The third bar shows the primary combinations of MLST clonal complexes for C. jejuni and C. coli, with red-labeled clonal complexes representing livestock-associated lineages, blue-labeled clonal complexes representing water and wildlife-associated lineages (Stabler et al., 2013), with the exception of the ST-61 clonal complex, which has been described as cattle (livestock)-associated (Kwan et al., 2008; Rotariu et al., 2009). The asterisks at ST-42 and ST-353 indicate that these clonal complexes may have a proportion of livestock-associated isolates. The association of some clonal complexes such as ST-464 was not reported previously and these are in black font. The lowercase letters indicate the approximate position of reference strains 81116 and M1 (a), 81-176 (b), RM1221 (c) and NCTC 11168 (d).
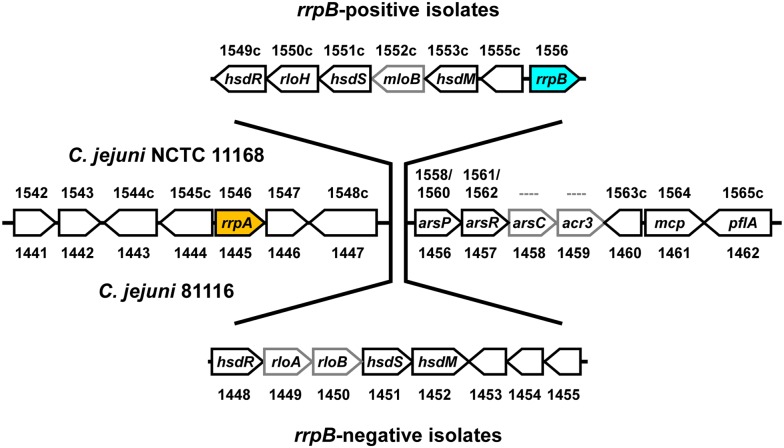
Further analysis of the C. jejuni and C. coli genome sequences investigated the distribution of genes flanking rrpA and rrpB and identified two conserved flanking regions with a more variable central region (Figure 2). The upstream flanking region contains rrpA whilst the downstream flanking region contains an arsenic resistance operon, an MCP-gene and a paralyzed flagella gene pflA. The hypervariable central region contains a type I Restriction-Modification (hsd) system. The NCTC 11168 version of this hypervariable central region contains rrpB whilst the 81116 version does not (Supplementary Table 3). The 81116 version of this hypervariable central region is representative of all the other C. jejuni ST strains analyzed in this study. Analysis of the C. coli genome sequences (which have primarily a structure very similar to 81116) has versions mostly without rrpA and rrpB.
FIGURE 2.
NCTC 11168 type (rrpB+ isolate) and 81116 type (rrpB- isolate) differ in a hypervariable region containing a type I Restriction-Modification (hsd) system. The genetic structure around rrpA is conserved in C. jejuni strains, however, rrpB appears to be part of a transferable hypervariable region linked with variation in the type I Restriction-Modification (hsd) system, suggesting a mechanism for the variable distribution of rrpB. The figure shows the C jejuni NCTC 11168 (rrpB+) and 81116 (rrpB-) genes and gene numbers (Supplementary Table 3).
Campylobacter jejuni rrpB- Wild-Type Strains Exhibit Increased Resistance to Oxidative Stress Compared to C. jejuni rrpB+ Wild-Type Strains
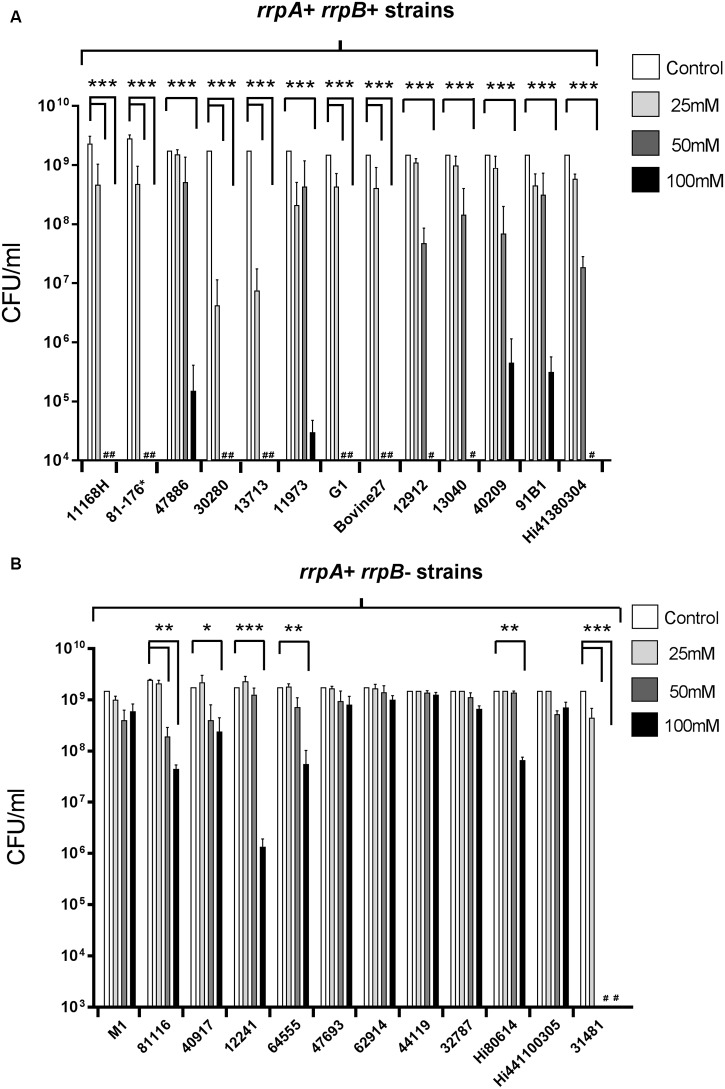
Characterization of the role of RrpA and RrpB in the C. jejuni 11168H wild-type strain has shown that both rrpA and rrpB mutants exhibit increased sensitivity to H2O2 when compared to the wild-type strain (Gundogdu et al., 2011, 2015). Based on the observed variation of rrpA and rrpB amongst C. jejuni isolates (Gundogdu et al., 2011), we utilized the Stabler et al. (2013) study to select 25 wild-type strains (Table 1) (13 rrpB+ and 12 rrpB- strains) for testing sensitivity toward H2O2 (Figure 3). rrpB- strains (Figure 3B) displayed significantly greater resistance to H2O2 when compared to rrpB+ strains (Figure 3A). The variation in the presence of rrpA and rrpB identified from the microarray data (Champion et al., 2005) for these 25 strains was also confirmed using PCR (data not shown).
FIGURE 3.
Effect of oxidative stress on the survival of C. jejuni rrpB+(A) and rrpB-(B) wild-type strains. rrpB+ strains are 11168H, 81-176, 47886, 30280, 13713, 11973, G1, Bovine27, 12912, 13040, 40209, 91B1, and Hi41380304. rrpB- strains are M1, 81116, 40917, 12241, 64555, 47693, 62914, 44119, 32787, Hi80614, Hi441100305, and 31481. These strains were selected from the Stabler et al. (2013) study (Table 1). C. jejuni strains were incubated with 25 mM, 50 or 100 mM H2O2 for 15 min at 37°C under microaerobic conditions. ∗For 81-176 denotes that this strain was not included in Stabler et al. (2013), but was selected here as a common laboratory strain. Bacterial survival was subsequently assessed. # Symbol indicates lack of growth. Asterisks denote a statistically significant difference (∗ = p < 0.05, ∗∗ = p < 0.01, ∗∗∗ = p < 0.001) between strains.
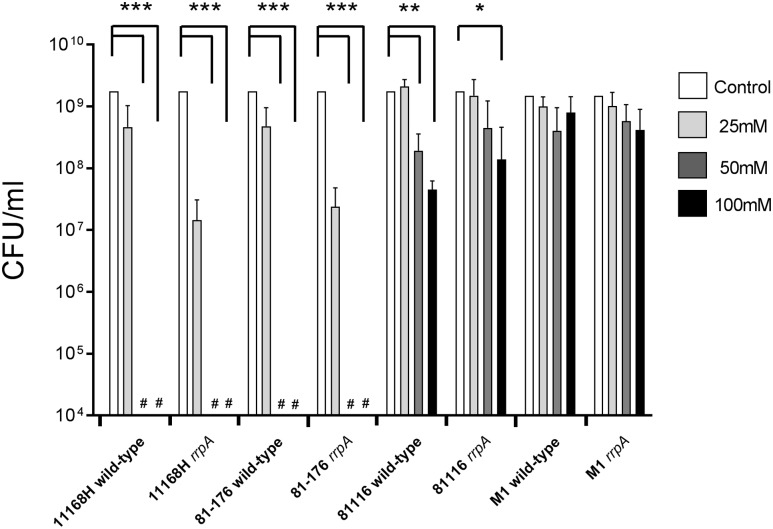
Mutation of rrpA Results in Increased Sensitivity to Peroxide Stress in Strains 11168H and 81-176 (rrpB+), But Not in Strains 81116 and M1 (rrpB-)
To further investigate the link between oxidative stress resistance and the varied distribution of rrpA and rrpB, rrpA and rrpB single mutants were constructed in the 11168H and 81-176 wild-type strains (rrpB+) and rrpA mutants were also constructed in the 81116 and M1 wild-type strains (rrpB-). The mutants obtained did not have any observable deficiency in motility or growth rate under standard conditions (data not shown). Compared to the respective wild-type strains, both the 11168H and 81-176 rrpA mutants exhibited an increased sensitivity to 25 mM H2O2, however, the 81116 and M1 rrpA mutants exhibited wild-type levels of survival (Figure 4). Neither the 11168H and 81-176 wild-type strains nor the respective rrpA mutants survived exposure to higher concentrations of H2O2 (50 or 100 mM), however, the 81116 and M1 wild-type strains and the respective rrpA mutants all exhibited only slightly increased sensitivity to these higher concentrations of H2O2 (Figure 4). 11168H and 81-176 rrpB mutants exhibited the same increased sensitivity to 25 mM H2O2 as the rrpA mutants (Figure 5), but did not survive the higher concentrations of H2O2 (data not shown). Cumene hydroperoxide and menadione stress assays were also performed on the 11168H, 81-176, 81116, M1 wild-type strains and the respective rrpA and rrpB mutants (Supplementary Figure 3). There were no significant differences in sensitivity to either cumene hydroperoxide or menadione between any of the wild-type strains and respective mutants, with the exception that the 81116 rrpA mutant appeared to be more resistant to cumene hydroperoxide stress than the 81116 wild-type strain.
FIGURE 4.
Effect of oxidative stress on the survival of C. jejuni wild-type strains 11168H, 81-176, 81116, and M1 and respective rrpA mutants. C. jejuni strains were incubated with 25, 50, or 100 mM H2O2 for 15 min at 37°C under microaerobic conditions. Bacterial survival was subsequently assessed. # Symbol indicates lack of growth. Asterisks denote a statistically significant difference (∗ = p < 0.05, ∗∗ = p < 0.01, ∗∗∗ = p < 0.001) between wild-type and mutant strains.
FIGURE 5.
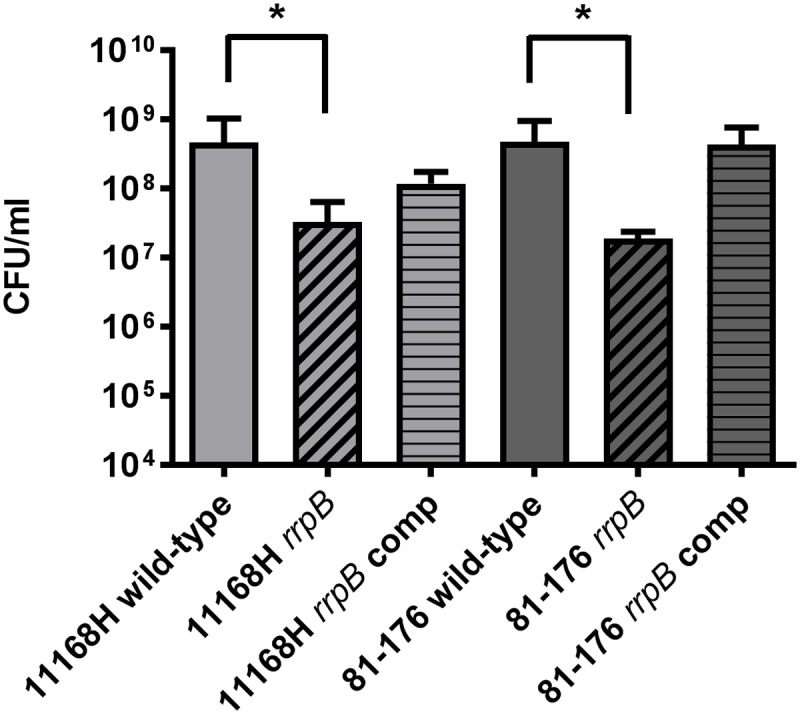
Effect of oxidative stress on the survival of C. jejuni wild-type strains 11168H and 81-176, rrpB mutants and rrpB complements. C. jejuni strains were incubated with 25 mM H2O2 for 15 min at 37°C under microaerobic conditions. Bacterial survival was subsequently assessed. Asterisks denote a statistically significant difference (∗ = p < 0.05) between wild-type and mutant strains.
Mutation of rrpA Reduces Growth under Aerobic Stress Conditions in rrpB+ Strains, But Not in rrpB- Strains
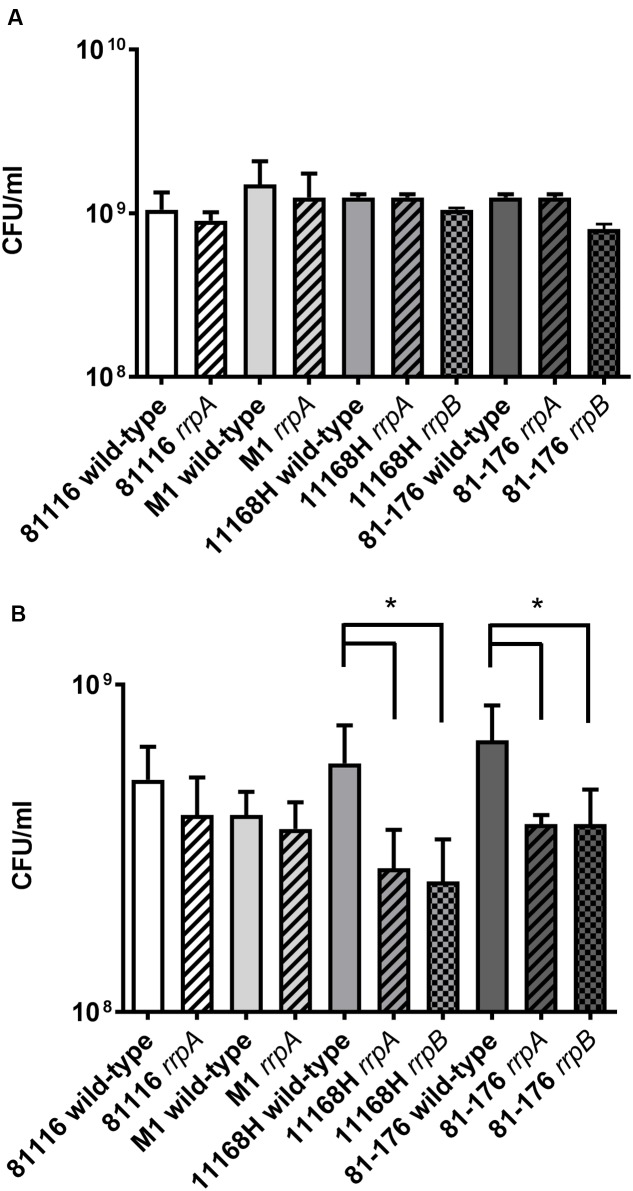
In C. jejuni 11168H, rrpA and rrpB mutants exhibit reduced growth under aerobic stress conditions (Gundogdu et al., 2011, 2015). The ability of rrpA and rrpB mutants to grow under aerobic stress conditions was compared to the respective four wild-type strains. No differences in bacterial growth kinetics (Supplementary Figure 4A) or in CFU counts after 16 h (Figure 6A) was observed under microaerobic conditions. However, differences were observed under aerobic conditions. All four rrpA mutants displayed significantly reduced growth kinetics during the logarithmic phase under aerobic conditions (Supplementary Figure 4B). However, compared to the respective wild-type strains, only the 11168H and 81-176 rrpA mutants exhibited a reduction in CFU counts after 16 h, whilst the 81116 and M1 rrpA mutants did not (Figure 6B).
FIGURE 6.
Growth assays on C. jejuni 11168H, 81-176, 81116 and M1 wild-type strains and respective rrpA and rrpB mutants. Brucella broth was pre-incubated at 37°C under microaerobic conditions for 24 h, then inoculated with a bacterial suspension to a final OD600 of 0.1 and grown for 16 h with shaking at 75 rpm at 37°C under either microaerobic conditions (A) or aerobic conditions (B). Bacterial CFU were also assessed at 16 h. Asterisks denote a statistically significant difference (∗ = p < 0.05) between wild-type and mutant strains.
Mutation of rrpA Results in Decreased Cytotoxicity in the Galleria mellonella Larvae Model of Infection
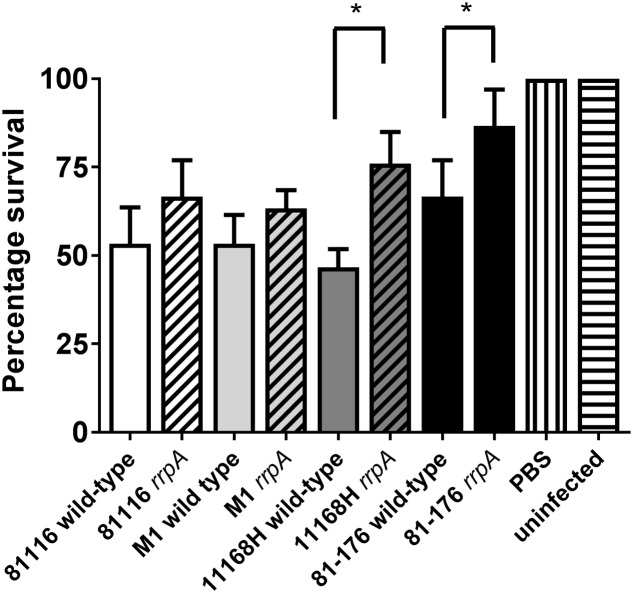
Galleria mellonella larvae have been used as a model to study infection with many different enteric pathogens including C. jejuni (Champion et al., 2009). Insect larvae possess specialized phagocytic cells, termed haemocytes (Bergin et al., 2005; Mylonakis et al., 2007). Haemocytes mimic the functions of phagocytic cells in mammals and are able to degrade bacterial pathogens as well as generate bactericidal compounds such as superoxide via a respiratory burst (Lavine and Strand, 2002; Bergin et al., 2005). Both 11168H rrpA and rrpB mutants have been shown to exhibit reduced cytotoxicity in this model of infection compared to the wild-type strain (Gundogdu et al., 2011, 2015). The cytotoxicity of these mutants in G. mellonella larvae compared to the respective wild-type strains was investigated. Infection with either the 11168H or 81-176 rrpA mutants resulted in a statistically significant decrease in cytotoxicity to G. mellonella larvae compared to infection with the respective wild-type strains (Figure 7). Infection with the 81116 or M1 rrpA mutants also resulted in a decrease in cytotoxicity to G. mellonella larvae compared to infection with the respective wild-type strains, although this decrease was not statistically significant (Figure 7).
FIGURE 7.
Galleria mellonella infection model on C. jejuni 11168H, 81-176, 81116, and M1 wild-type strains and respective rrpA and rrpB mutants. G. mellonella larvae were injected with a 10 μl inoculum of a 24 h C. jejuni culture diluted to OD600 0.1 by micro-injection in the right foremost leg, giving an infectious dose of approximately 106 CFU. Larvae were incubated at 37°C with survival and appearance recorded at 24 h intervals. PBS and no injection controls were used. For each experiment, 10 G. mellonella larvae were infected and experiments were repeated in triplicate. Asterisks denote a statistically significant difference (∗ = p < 0.05) between wild-type and mutant strains.
Mutation of rrpA Results in Increased Biofilm Formation
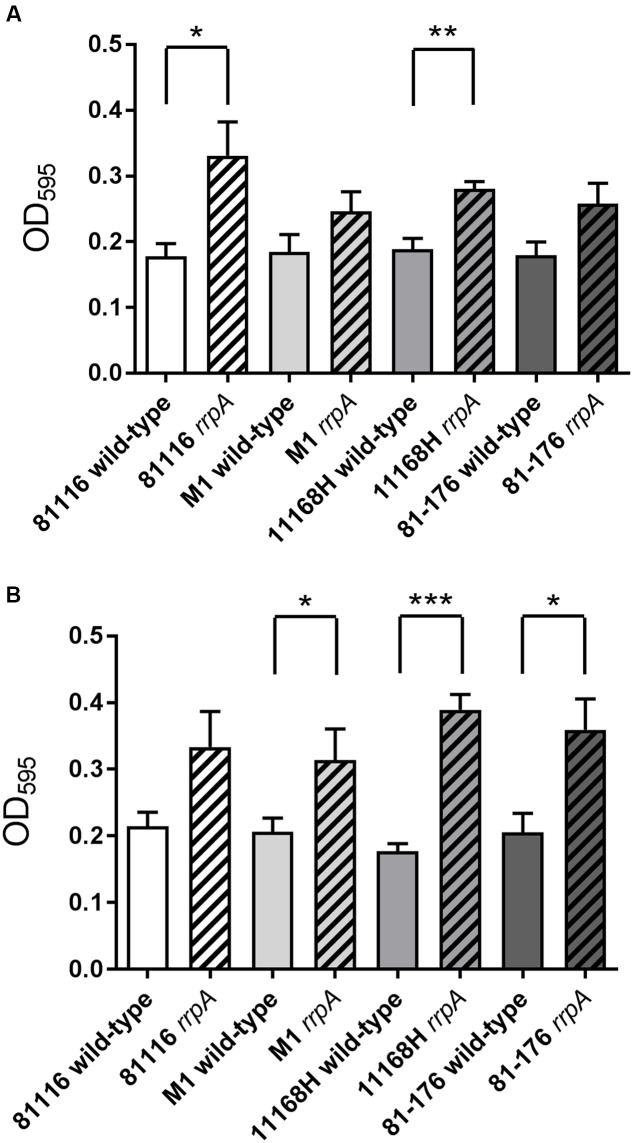
Several studies have demonstrated that C. jejuni exhibits increased biofilm formation under increased oxidative stress conditions (Fields and Thompson, 2008; Reuter et al., 2010). Previously C. jejuni 11168H rrpA and rrpB mutants exhibited an enhanced ability to form biofilms under both aerobic and microaerobic conditions (Gundogdu et al., 2015). All four rrpA mutants exhibited an increase in biofilm formation under both aerobic (Figure 8A) and microaerobic (Figure 8B) conditions after 72 h compared to the respective wild-type strains, which was statistically significant for the 11168H and 81116 rrpA mutants under aerobic conditions and for the 11168H, 81-176, and M1 rrpA mutants under microaerobic conditions. In addition both the 11168H rrpB mutant (Gundogdu et al., 2015) and 81-176 rrpB mutant (data not shown) exhibit increased biofilm formation under aerobic and microaerobic conditions when compared to the respective wild-type strains.
FIGURE 8.
Biofilm assay on C. jejuni 11168H, 81-176, 81116, and M1 wild-type strains and respective rrpA and rrpB mutants. C. jejuni 11168H wild-type strain and mutants were grown for 72 h under aerobic (A) and microaerobic (B) growth conditions at 37°C without shaking, rinsed three times with PBS, followed by crystal violet staining. Asterisks denote a statistically significant difference (∗ = p < 0.05, ∗∗ = p < 0.01, ∗∗∗ = p < 0.001) between strains.
Discussion
Campylobacter jejuni will be exposed to ROS both during colonization or infection of a host and in the environment as well as during the course of normal bacterial metabolism. It remains a conundrum as to how this microaerophilic pathogen is so widely dispersed, highly prevalent and able to survive in the ambient environment. C. jejuni contains a number of different mechanisms for counteracting the effects of oxidative stress and the control of the C. jejuni oxidative stress response is complex involving multiple inter-linked levels of regulation (Atack and Kelly, 2009). The re-annotation of the C. jejuni NCTC 11168 genome sequence (Gundogdu et al., 2007) identified both RrpA and RrpB as putative MarR-type transcriptional regulators which were subsequently shown to be involved in the C. jejuni peroxide and aerobic stress response (Gundogdu et al., 2011, 2015). All the other regulators of the C. jejuni oxidative stress response, such as PerR, Fur, CosR, CsrA, CprRS, and RacRS, are all conserved (>99.8%) amongst all the C. jejuni and C. coli wild-type strains in this study (van Vliet et al., 1999; Atack and Kelly, 2009; Palyada et al., 2009; Hwang et al., 2012). Analysis of the distribution of rrpA and rrpB in 3,746 C. jejuni and 486 C. coli genomes from public databases confirmed that the vast majority of C. jejuni strains contain rrpA, whilst the presence of rrpB is more restricted, with the distribution linked to MLST clonal complex. The majority of C. jejuni strains that contain both rrpA and rrpB are from ST-21 and ST-61. ST-21 strains are often associated with human infections (Sheppard et al., 2009) and ST-61 strains with infections of livestock (Kwan et al., 2008; Rotariu et al., 2009).
Analysis of the distribution of rrpA and rrpB from the microarray data study of 270 C. jejuni strains (Stabler et al., 2013) identified water and wildlife strains predominantly as rrpB-, whereas livestock-associated strains were predominantly rrpB+ (Supplementary Figure 1; Supplementary Table 1). This distribution of rrpA and rrpB was also reflected in the analysis of the whole genome sequence data (Figure 1) where red-labeled clonal complexes represent livestock-associated lineages and blue-labeled clonal complexes represent water and wildlife-associated lineages. It is, however, not always clear whether each clonal complex is associated with a single host as clonal complexes are regularly isolated from multiple animal species and thus attribution to a single host reservoir is difficult using MLST data alone (Dearlove et al., 2016). One example is the ST-48 complex that has been isolated from humans, cattle and sand from beaches (Dingle et al., 2002). Another example is ST-257 and ST-61 that have been isolated from chicken and ruminants, yet ST-257 is designated as a water and wildlife-associated lineage, whereas ST-61 is denoted as a livestock-associated lineage (Sheppard et al., 2011). These livestock and water and wildlife descriptions may be somewhat generalized and investigating the properties of individual STs will be more important to understand the source. ST-21, ST-206, and ST-48 may form a “complex group” of related genotypes that are widely distributed, perhaps reflecting the “generalist” ability to colonize a wide range of hosts (Dingle et al., 2002; Sheppard et al., 2011).
The location of rrpA and rrpB close to a type I Restriction-Modification (hsd) system suggests an explanation as to why rrpA is present in almost all C. jejuni strains, whilst the presence of rrpB is restricted and appears to correlate with MLST clonal complexes. Restriction-modification (R-M) systems are ubiquitous in the bacterial world and provide a defense against foreign DNA and bacteriophages (Wilson and Murray, 1991). Foreign DNA is cleaved by endonucleases but host DNA avoids damage due to methylation. R-M systems have been classified into four distinct groups, type I, type II, type III, and type IV. A type I R-M locus was identified in the genome sequence of NCTC 11168 (Cj1549–Cj1553) (Parkhill et al., 2000). The type I enzyme is a bi-functional, multi-subunit complex consisting of HsdR, HsdM, and HsdS. Further analysis of the type I R-M system from 73 C. jejuni strains (including NCTC 11168, 81-176, and 81116) assigned some hsd systems to the classical type IC family but also identified two additional type I R-M families, termed type IAB and type IF (Miller et al., 2005). 81116 contains a type IAB hsd system, whilst both NCTC 11168 and 81-176 contain a type IF hsd system. This study also found evidence for extensive rearrangements within the C. jejuni hsd loci of the same family but suggested that the low sequence similarity between the IC, IAB, and IF families would make inter-family recombination events unlikely. The NCTC 11168 version of the hypervariable central region identified in this study, which contains the type IF hsd system and rrpB, represents the majority of ST-21, ST-61, ST-42, and ST-353 strains. 81-176 also contains the type IF hsd system and rrpB. However, the 81116 version of this hypervariable central region, which represents the majority of all the other C. jejuni ST strains analyzed in this study, contains a type IAB hsd system and does not contain rrpB. The lack of recombination between C. jejuni strains containing the type IF hsd system and rrpB with other C. jejuni strains containing a different family of type I hsd system could be the explanation for the restricted distribution of rrpB amongst C. jejuni strains. This region of the C. jejuni genome has previously been identified as hypervariable region 14, one of 16 hypervariable regions (Taboada et al., 2004). The presence of intervening ORFs between hsdR and hsdS (referred to as rlo genes for R-linked ORF) and also between hsdS and hsdM (referred to as mlo genes for M-linked ORFs) are a distinct feature of the C. jejuni hsd loci (Miller et al., 2005). H. pylori ModH from the type III R-M system has been shown to have a regulatory role (Srikhanta et al., 2011) suggesting that R-M systems may also have a role in the regulation of virulence gene expression (Vasu and Nagaraja, 2013). Recently the same hypervariable region in C. coli has been shown to contain a novel streptomycin resistance gene (Olkkola et al., 2016). The genomics-based screening also showed that there are different alleles of RrpA and RrpB (Supplementary Figure 2) leading to a N-terminal truncation, a C-terminal truncation or both. The truncated version of RrpB was predominantly observed in isolates of the ST-353 clonal complex, while truncated versions of RrpA were rare in most clonal complexes, but dominant in ST-607 isolates (Supplementary Table 2). Future studies could utilize these truncated versions to test for the role of the N-terminal and C-terminal regions in functionality of RrpA and RrpB.
Campylobacter jejuni rrpB- strains exhibit a pattern of greater resistance to peroxide stress when compared to rrpB+ strains. This pattern was the same when comparing the 11168H, 81-176, 81116, and M1 rrpA mutants against the respective wild-type strain where rrpA mutants in rrpB- strains were more resistant to peroxide stress compared to rrpA mutants in rrpB+ strains. In addition, when comparing bacterial growth of 11168H, 81-176, 81116, and M1 rrpA mutants against the respective wild-type strain under aerobic conditions, all four rrpA mutants displayed a reduced growth rate during the logarithmic phase. Both the 11168H and 81-176 rrpA mutants exhibited lower CFUs at 16 h compared to the respective wild-type strains. Even though both 81116 and M1 rrpA mutants also displayed slower growth rates compared to the respective wild-type strains when comparing respective CFU counts at 16 h, these differences were not statistically significant. The general pattern that rrpB+ strains exhibit increased sensitivity to peroxide and aerobic stresses is rather counterintuitive. Possibly having only a single Rrp regulator is more efficient in responding to such stresses. Certainly rrpA is the most conserved amongst C. jejuni strains. Even though the oxidative stress assays were performed on a relatively small number of selected strains, it is interesting to speculate why livestock-associated strains such as NCTC 11168 (ST-21) would be more sensitive to peroxide and aerobic stress, whilst water and wildlife-associated strains such as 81116 (ST-283) and M1 (ST-45) would be more resistance to such stresses is difficult to explain. One possible hypothesis is that the ability to survive aerobic stress is more important for the latter strains. The variation in the presence of rrpA and rrpB between different wild-type strains may play an important role in the ability of C. jejuni to adapt and survive in different environmental niches.
Infection with rrpA mutants from both rrpB+ and rrpB- strains resulted in an increase in survival of G. mellonella larvae compared to infection with the respective wild-type strains. We have previously demonstrated that infection of G. mellonella with the 11168H rrpA mutant leads to increased survival of the G. mellonella larvae (Gundogdu et al., 2015). Here we demonstrate that the 81-176 rrpA mutant (rrpB+) also exhibits a similar statistically significant reduction in larval cytotoxicity. Both 81116 and M1 rrpA mutants (rrpB-) exhibit reduced larval cytotoxicity, but not to a significant degree. This suggests the rrpA mutants (like the rrpB mutants) are more susceptible to the host immune mechanisms resulting in reduced bacterial survival within G. mellonella. This also suggests that RrpA may in fact play a role in the oxidative stress response of rrpB- strains such as 81116 and M1, but at a different level compared to rrpB+ strains, such as 11168H and 81-176. Certainly the enhanced resistance of the 81116 and M1 rrpA mutants to peroxide stress compared to the 11168H and 81-176 rrpA mutants is not reflected by increased virulence in the G. mellonella larvae infection model.
Campylobacter jejuni forms biofilms (Joshua et al., 2006; Gundogdu et al., 2011) and this may be an important factor in the survival of C. jejuni both within hosts and in the environment. Though our understanding of the specific mechanisms underlying biofilm formation in C. jejuni is still limited (Svensson et al., 2008), C. jejuni lacks the classical two component regulatory systems involved in biofilm formation that are present in other bacteria such as GacSA in Pseudomonas aeruginosa (Parkins et al., 2001). Biofilm formation has been linked to responses to oxidative and aerobic stress as C. jejuni biofilm formation is increased under aerobic conditions (Reuter et al., 2010) and mutation of genes encoding oxidative stress response proteins results in changes in biofilm formation (Oh and Jeon, 2014; Gundogdu et al., 2015). Analysis of biofilm formation demonstrated that rrpA mutants from both rrpB+ and rrpB- strains exhibited an increased biofilm phenotype compared to the respective wild-type strain under both microaerobic and aerobic conditions. Again the enhanced resistance of the 81116 and M1 rrpA mutants to peroxide stress compared to the 11168H and 81-176 rrpA mutants is not reflected by a decrease in biofilm formation.
The basis of C. jejuni survival is dependent upon the ability to sense and respond to the different environments encountered within hosts and in the environment. In this study we identified bioinformatically that over 99% of C. jejuni strains contain rrpA, whilst rrpB is restricted and appears to correlate with livestock-associated MLST clonal complexes. There exists a conserved genetic structure for rrpA, whilst rrpB seems to be part of a transferable hypervariable region linked with variation in the type I R-M (hsd) system, giving an explanation for the more restricted distribution of rrpB amongst C. jejuni strains. rrpB- strains possess an increased level of resistance to peroxide and aerobic stress compared to rrpB+ strains. So whilst all the other oxidative stress response regulators such as PerR, Fur, CosR, CsrA, CprRS, RacRS, and RrpA appear to be conserved in C. jejuni, variation in the presence of RrpB between different wild-type strains may play an important role for altered oxidative stress responses through the concerted actions of these multiple regulators in this microaerophilic pathogen. This highlights the potential of genetic variation in the natural population in the adaptation to different environmental niches.
Author Contributions
OG, DdS, BW, and ND designed this study. OG, DdS, BM, and AE performed all the lab-based experimental work. OG, DdS, and AvV performed all the bioinformatic analysis. OG, DdS, BW, AvV, and ND wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
DdS is funded through a Doctorate Scholarship from CNPq in Brazil. We thank Sarah Leir and Naomi Henderson for technical assistance. This publication made use of the PubMLST website (http://pubmlst.org/) developed by Keith Jolley and sited at the University of Oxford. The development of that website was funded by the Wellcome Trust.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.02117/full#supplementary-material
References
- Atack J. M., Kelly D. J. (2009). Oxidative stress in Campylobacter jejuni: responses, resistance and regulation. Future Microbiol. 4 677–690. 10.2217/fmb.09.44 [DOI] [PubMed] [Google Scholar]
- Bergin D., Reeves E. P., Renwick J., Wientjes F. B., Kavanagh K. (2005). Superoxide production in Galleria mellonella hemocytes: identification of proteins homologous to the NADPH oxidase complex of human neutrophils. Infect. Immun. 73 4161–4170. 10.1128/IAI.73.7.4161-4170.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown H. L., Reuter M., Hanman K., Betts R. P., Van Vliet A. H. (2015). Prevention of biofilm formation and removal of existing biofilms by extracellular DNases of Campylobacter jejuni. PLoS ONE 10:e0121680 10.1371/journal.pone.0121680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne C. M., Clyne M., Bourke B. (2007). Campylobacter jejuni adhere to and invade chicken intestinal epithelial cells in vitro. Microbiology 153 561–569. 10.1099/mic.0.2006/000711-0 [DOI] [PubMed] [Google Scholar]
- Champion O. L., Cooper I. A., James S. L., Ford D., Karlyshev A., Wren B. W., et al. (2009). Galleria mellonella as an alternative infection model for Yersinia pseudotuberculosis. Microbiology 155 1516–1522. 10.1099/mic.0.026823-0 [DOI] [PubMed] [Google Scholar]
- Champion O. L., Gaunt M. W., Gundogdu O., Elmi A., Witney A. A., Hinds J., et al. (2005). Comparative phylogenomics of the food-borne pathogen Campylobacter jejuni reveals genetic markers predictive of infection source. Proc. Natl. Acad. Sci. U.S.A 102 16043–16048. 10.1073/pnas.0503252102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion O. L., Karlyshev A. V., Senior N. J., Woodward M., La Ragione R., Howard S. L., et al. (2010). Insect infection model for Campylobacter jejuni reveals that O-methyl phosphoramidate has insecticidal activity. J. Infect. Dis. 201 776–782. 10.1086/650494 [DOI] [PubMed] [Google Scholar]
- Cody A. J., Mccarthy N. D., Jansen Van Rensburg M., Isinkaye T., Bentley S. D., Parkhill J., et al. (2013). Real-time genomic epidemiological evaluation of human Campylobacter isolates by use of whole-genome multilocus sequence typing. J. Clin. Microbiol. 51 2526–2534. 10.1128/JCM.00066-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Autreaux B., Toledano M. B. (2007). ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 8 813–824. 10.1038/nrm2256 [DOI] [PubMed] [Google Scholar]
- Dearlove B. L., Cody A. J., Pascoe B., Meric G., Wilson D. J., Sheppard S. K. (2016). Rapid host switching in generalist Campylobacter strains erodes the signal for tracing human infections. ISME J. 10 721–729. 10.1038/ismej.2015.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingle K. E., Colles F. M., Ure R., Wagenaar J. A., Duim B., Bolton F. J., et al. (2002). Molecular characterization of Campylobacter jejuni clones: a basis for epidemiologic investigation. Emerg. Infect. Dis. 8 949–955. 10.3201/eid0809.020122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi R., Nothaft H., Garber J., Xin Kin L., Stahl M., Flint A., et al. (2016). L-fucose influences chemotaxis and biofilm formation in Campylobacter jejuni. Mol. Microbiol. 101 575–589. 10.1111/mmi.13409 [DOI] [PubMed] [Google Scholar]
- Fang F. C. (2004). Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat. Rev. Microbiol. 2 820–832. 10.1038/nrmicro1004 [DOI] [PubMed] [Google Scholar]
- Fields J. A., Thompson S. A. (2008). Campylobacter jejuni CsrA mediates oxidative stress responses, biofilm formation, and host cell invasion. J. Bacteriol. 190 3411–3416. 10.1128/JB.01928-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friis C., Wassenaar T. M., Javed M. A., Snipen L., Lagesen K., Hallin P. F., et al. (2010). Genomic characterization of Campylobacter jejuni strain M1. PLoS ONE 5:e12253 10.1371/journal.pone.0012253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundogdu O., Bentley S. D., Holden M. T., Parkhill J., Dorrell N., Wren B. W. (2007). Re-annotation and re-analysis of the Campylobacter jejuni NCTC11168 genome sequence. BMC Genomics 8:162 10.1186/1471-2164-8-162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundogdu O., Da Silva D. T., Mohammad B., Elmi A., Mills D. C., Wren B. W., et al. (2015). The Campylobacter jejuni MarR-like transcriptional regulators RrpA and RrpB both influence bacterial responses to oxidative and aerobic stresses. Front. Microbiol. 6:724 10.3389/fmicb.2015.00724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundogdu O., Mills D. C., Elmi A., Martin M. J., Wren B. W., Dorrell N. (2011). The Campylobacter jejuni transcriptional regulator Cj1556 plays a role in the oxidative and aerobic stress response and is important for bacterial survival in vivo. J. Bacteriol. 193 4238–4249. 10.1128/JB.05189-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handley R. A., Mulholland F., Reuter M., Ramachandran V. K., Musk H., Clissold L., et al. (2015). PerR controls oxidative stress defence and aerotolerance but not motility-associated phenotypes of Campylobacter jejuni. Microbiology 161 1524–1536. 10.1099/mic.0.000109 [DOI] [PubMed] [Google Scholar]
- Hwang S., Kim M., Ryu S., Jeon B. (2011). Regulation of oxidative stress response by CosR, an essential response regulator in Campylobacter jejuni. PLoS ONE 6:e22300 10.1371/journal.pone.0022300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S., Zhang Q., Ryu S., Jeon B. (2012). Transcriptional regulation of the CmeABC multidrug efflux pump and the KatA catalase by CosR in Campylobacter jejuni. J. Bacteriol. 194 6883–6891. 10.1128/JB.01636-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay J. A. (2003). Pathways of oxidative damage. Annu. Rev. Microbiol. 57 395–418. 10.1146/annurev.micro.57.030502.090938 [DOI] [PubMed] [Google Scholar]
- Jolley K. A., Maiden M. C. (2010). BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595 10.1186/1471-2105-11-595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. A., Marston K. L., Woodall C. A., Maskell D. J., Linton D., Karlyshev A. V., et al. (2004). Adaptation of Campylobacter jejuni NCTC11168 to high-level colonization of the avian gastrointestinal tract. Infect. Immun. 72 3769–3776. 10.1128/IAI.72.7.3769-3776.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshua G. W., Guthrie-Irons C., Karlyshev A. V., Wren B. W. (2006). Biofilm formation in Campylobacter jejuni. Microbiology 152 387–396. 10.1099/mic.0.28358-0 [DOI] [PubMed] [Google Scholar]
- Karlyshev A. V., Linton D., Gregson N. A., Wren B. W. (2002). A novel paralogous gene family involved in phase-variable flagella-mediated motility in Campylobacter jejuni. Microbiology 148 473–480. 10.1099/00221287-148-2-473 [DOI] [PubMed] [Google Scholar]
- Kim J. C., Oh E., Kim J., Jeon B. (2015). Regulation of oxidative stress resistance in Campylobacter jejuni, a microaerophilic foodborne pathogen. Front. Microbiol. 6:751 10.3389/fmicb.2015.00751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korlath J. A., Osterholm M. T., Judy L. A., Forfang J. C., Robinson R. A. (1985). A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J. Infect. Dis. 152 592–596. 10.1093/infdis/152.3.592 [DOI] [PubMed] [Google Scholar]
- Kruczkiewicz P., Mutschall S., Barker D., Thomas J. E., Domselaar G. V. H., Gannon V., et al. (2013). “MIST: a tool for rapid in silico generation of molecular data from bacterial genome sequences,” in Proceedings of the 4th International Conference on Bioinformatics Models, Methods and Algorithms, Barcelona, 316–323. [Google Scholar]
- Kwan P. S., Birtles A., Bolton F. J., French N. P., Robinson S. E., Newbold L. S., et al. (2008). Longitudinal study of the molecular epidemiology of Campylobacter jejuni in cattle on dairy farms. Appl. Environ. Microbiol. 74 3626–3633. 10.1128/AEM.01669-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavine M. D., Strand M. R. (2002). Insect hemocytes and their role in immunity. Insect Biochem. Mol. Biol. 32 1295–1309. 10.1016/S0965-1748(02)00092-9 [DOI] [PubMed] [Google Scholar]
- Miller W. G., Pearson B. M., Wells J. M., Parker C. T., Kapitonov V. V., Mandrell R. E. (2005). Diversity within the Campylobacter jejuni type I restriction-modification loci. Microbiology 151 337–351. 10.1099/mic.0.27327-0 [DOI] [PubMed] [Google Scholar]
- Mylonakis E., Casadevall A., Ausubel F. M. (2007). Exploiting amoeboid and non-vertebrate animal model systems to study the virulence of human pathogenic fungi. PLoS Pathog. 3:e101 10.1371/journal.ppat.0030101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Jeon B. (2014). Role of alkyl hydroperoxide reductase (AhpC) in the biofilm formation of Campylobacter jejuni. PLoS ONE 9:e87312 10.1371/journal.pone.0087312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olkkola S., Culebro A., Juntunen P., Hanninen M. L., Rossi M. (2016). Functional genomics in Campylobacter coli identified a novel streptomycin resistance gene located in a hypervariable genomic region. Microbiology 162 1157–1166. 10.1099/mic.0.000304 [DOI] [PubMed] [Google Scholar]
- Page A. J., Cummins C. A., Hunt M., Wong V. K., Reuter S., Holden M. T., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31 3691–3693. 10.1093/bioinformatics/btv421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palyada K., Sun Y. Q., Flint A., Butcher J., Naikare H., Stintzi A. (2009). Characterization of the oxidative stress stimulon and PerR regulon of Campylobacter jejuni. BMC Genomics 10:481 10.1186/1471-2164-10-481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J., Wren B. W., Mungall K., Ketley J. M., Churcher C., Basham D., et al. (2000). The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403 665–668. 10.1038/35001088 [DOI] [PubMed] [Google Scholar]
- Parkins M. D., Ceri H., Storey D. G. (2001). Pseudomonas aeruginosa GacA, a factor in multihost virulence, is also essential for biofilm formation. Mol. Microbiol. 40 1215–1226. 10.1046/j.1365-2958.2001.02469.x [DOI] [PubMed] [Google Scholar]
- Pearson B. M., Louwen R., Van Baarlen P., Van Vliet A. H. (2015). Differential Distribution of Type II CRISPR-Cas systems in agricultural and nonagricultural Campylobacter coli and Campylobacter jejuni isolates correlates with lack of shared environments. Genome Biol. Evol. 7 2663–2679. 10.1093/gbe/evv174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M., Mallett A., Pearson B. M., Van Vliet A. H. (2010). Biofilm formation by Campylobacter jejuni is increased under aerobic conditions. Appl. Environ. Microbiol. 76 2122–2128. 10.1128/AEM.01878-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotariu O., Dallas J. F., Ogden I. D., Macrae M., Sheppard S. K., Maiden M. C., et al. (2009). Spatiotemporal homogeneity of Campylobacter subtypes from cattle and sheep across northeastern and southwestern Scotland. Appl. Environ. Microbiol. 75 6275–6281. 10.1128/AEM.00499-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Palacios G. M. (2007). The health burden of Campylobacter infection and the impact of antimicrobial resistance: playing chicken. Clin. Infect. Dis 44 701–703. 10.1086/509936 [DOI] [PubMed] [Google Scholar]
- Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30 2068–2069. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- Sheppard S. K., Colles F. M., Mccarthy N. D., Strachan N. J., Ogden I. D., Forbes K. J., et al. (2011). Niche segregation and genetic structure of Campylobacter jejuni populations from wild and agricultural host species. Mol. Ecol. 20 3484–3490. 10.1111/j.1365-294X.2011.05179.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard S. K., Dallas J. F., Strachan N. J., Macrae M., Mccarthy N. D., Wilson D. J., et al. (2009). Campylobacter genotyping to determine the source of human infection. Clin. Infect. Dis 48 1072–1078. 10.1086/597402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikhanta Y. N., Gorrell R. J., Steen J. A., Gawthorne J. A., Kwok T., Grimmond S. M., et al. (2011). Phasevarion mediated epigenetic gene regulation in Helicobacter pylori. PLoS ONE 6:e27569 10.1371/journal.pone.0027569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabler R. A., Larsson J. T., Al-Jaberi S., Nielsen E. M., Kay E., Tam C. C., et al. (2013). Characterization of water and wildlife strains as a subgroup of Campylobacter jejuni using DNA microarrays. Environ. Microbiol. 15 2371–2383. 10.1111/1462-2920.12111 [DOI] [PubMed] [Google Scholar]
- Svensson S. L., Frirdich E., Gaynor E. C. (2008). “Survival strategies of Campylobacter jejuni: stress responses, the viable but nonculturable state, and biofilms,” in Campylobacter, 3rd Edn, eds Nachmkin I., Szymanski C. M., Blaser M. J. (Washington, DC: ASM Press; ), 571–590. [Google Scholar]
- Taboada E. N., Acedillo R. R., Carrillo C. D., Findlay W. A., Medeiros D. T., Mykytczuk O. L., et al. (2004). Large-scale comparative genomics meta-analysis of Campylobacter jejuni isolates reveals low level of genome plasticity. J. Clin. Microbiol. 42 4566–4576. 10.1128/jcm.42.10.4566-4576.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu-Cuot P., Gerbaud G., Lambert T., Courvalin P. (1985). In vivo transfer of genetic information between gram-positive and gram-negative bacteria. EMBO J. 4 3583–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vliet A. H., Baillon M. L., Penn C. W., Ketley J. M. (1999). Campylobacter jejuni contains two fur homologs: characterization of iron-responsive regulation of peroxide stress defense genes by the PerR repressor. J. Bacteriol. 181 6371–6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vliet A. H., Kusters J. G. (2015). Use of alignment-free phylogenetics for rapid genome sequence-based typing of Helicobacter pylori virulence markers and antibiotic susceptibility. J. Clin. Microbiol. 53 2877–2888. 10.1128/JCM.01357-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vliet A. H., Rock J. D., Madeleine L. N., Ketley J. M. (2000). The iron-responsive regulator Fur of Campylobacter jejuni is expressed from two separate promoters. FEMS Microbiol. Lett. 188 115–118. 10.1111/j.1574-6968.2000.tb09180.x [DOI] [PubMed] [Google Scholar]
- van Vliet A. H., Wooldridge K. G., Ketley J. M. (1998). Iron-responsive gene regulation in a Campylobacter jejuni fur mutant. J. Bacteriol. 180 5291–5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasu K., Nagaraja V. (2013). Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol. Mol. Biol. Rev. 77 53–72. 10.1128/MMBR.00044-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenaar T. M., Bleumink-Pluym N. M., Van Der Zeijst B. A. (1991). Inactivation of Campylobacter jejuni flagellin genes by homologous recombination demonstrates that flaA but not flaB is required for invasion. EMBO J. 10 2055–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson G. G., Murray N. E. (1991). Restriction and modification systems. Annu. Rev. Genet. 25 585–627. 10.1146/annurev.ge.25.120191.003101 [DOI] [PubMed] [Google Scholar]
- Young K. T., Davis L. M., Dirita V. J. (2007). Campylobacter jejuni: molecular biology and pathogenesis. Nat. Rev. Microbiol. 5 665–679. 10.1038/nrmicro1718 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.