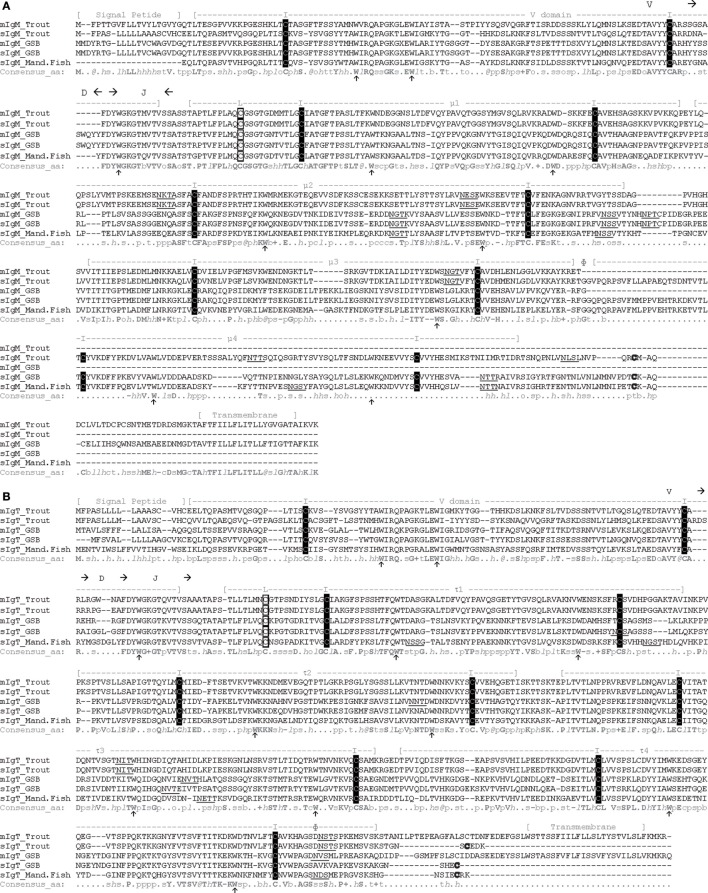
Figure 3.
Gilthead sea bream (GSB) soluble and membrane IgM and IgT are well conserved and have standard residues and structure. Amino acid sequence alignment of the soluble and membrane forms of IgM (A) and IgT (B) from rainbow trout, GSB, and mandarin fish (Mand.Fish) performed with PROMALS3D. The signal peptide, transmembrane region, and the stretches of amino acids predicted to constitute the VDJ segment and the variable or the different constant Ig domains (μ and τ) are depicted above the sequences. The alternative splicing region is signaled with Ф above the sequences. Conserved cysteine residues in black squares marked with I are the ones expected to form the intra-chain disulfide bonds, while cysteine residues in white squares marked with L are expected to constitute the disulfide bond to link the light chains to form the complete Ig molecule. Other conserved cysteine residues (bold) can be found at the C-terminal of all soluble Ig molecules. Arrows below the sequence point to the conserved tryptophan residues that help pack the structure. Underlined residues are the predicted N-glycosylation sites. Consensus amino acid (aa) symbols at the bottom of the alignment are: highly conserved aa, bold and uppercase letters; l, aliphatic; @, aromatic; h, hydrophobic; o, alcohol; p, polar residues; t, tiny; s, small; b, bulky residues; +, positively charged; −, negatively charged; c, charged. The GenBank accession numbers of the sequences used in this alignment are: trout: mIgM AAA56663, sIgM AAW66972, mIgT AAW66979, and sIgT AY870263; mandarin fish: sigm AAQ14863 and sIgT DQ016660; GSB: migm KX599199, sIgM JQ811851, mIgT KX599201, and sIgT KX599200.