Figure 2.
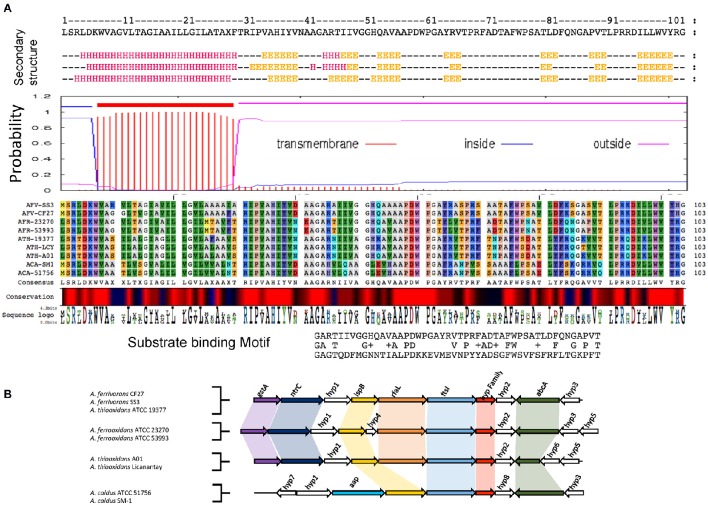
Example of functional prediction based on multiple bioinformatics and genome-based evidence for members of family II. (A) Bioinformatics analysis of members of family II based on secondary structure prediction, hydrophobicity profiles and transmembrane segments prediction, multiple alignments and conservation profiles for the generation of consensus and profile sequences and their comparison to specific substrate binding protein profiles found in public databases. (B) Genomic context analysis of members of family II including functional annotations of the closest neighborhood genes for functional association. gstA, Glutathione S-transferase; ntrC, Nitrogen assimilation regulatory protein; ispB, Octaprenyl-diphosphate synthase; rfaL, O-antigen ligase; ftsI, Cell division protein; Hyp family II, Hypothetical protein; abcA, ABC transporter A family; app, Amino acid permease; Hyp (1–8), Hypothetical protein (1–8). Table 2 provides a complete overview of the predicted properties from amino acid sequences for member of the five families.